Title: Imaging with Mass Spectrometry of Bacteria on the Exoskeleton of Fungus-Growing Ants
Authors: Erin Gemperline, Heidi A. Horn, Kellen Delaney, Cameron R. Currie, Lingjun Li
Year: 2017
Journal: ACS Chemical Biology
https://doi.org/10.1021/acschembio.7b00038
One of the most iconic examples of a microorganism-derived natural product is a drug familiar to all: penicillin. This classic antibiotic was first discovered when its parent mold invaded a culture of bacteria, inhibiting bacterial growth within the vicinity of the mold. The success of penicillin as a commercial antibiotic has prompted scientists to search for similar natural compounds in a variety of different organisms. Common methods for discovery of these compounds rely on simplistic “pairwise” interactions between a set of microbes (e.g. an organism responds to a pathogen by producing an antibiotic). The reality of these microbial interactions is usually much more complex, often involving multi-species interactions within a host organism. One can think of this as a small-scale battle between these different creatures, with some allied together to fight off a single foe!
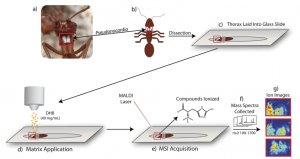
As a result of these miniature battles, more advanced methods to discover antibiotics have evolved. One method involves taking thin slices of these host species and analyzing the spatial distribution of chemicals inside the body of a host using mass spectrometry. However, although these techniques provide a wealth of information in the form of a chemical “map”, they are time-consuming and limited to 2D samples. The authors of this paper wished to move from 2D to 3D space, creating a similar map across the surface of an entire organism! They present a novel method capable of identifying specific natural products formed during interactions between members of a four-membered symbiotic relationship through mass spectrometry analysis of an entire ant.
The players in this symbiosis are: 1) the ant host, 2) a fungal food source, 3) the “good” bacteria Pseudonocardia, and 4) the pathogenic “bad” bacteria Escovopsis. The host ant cultivates its fungal food source and harbors Pseudonocardia (an ally) to ward off infestation of its food source by Escovopsis (an attacking foe!). The authors in the paper have cleverly adapted traditional mass spectrometry techniques to image the production of secondary metabolites formed on the chest of the ant itself.
This innovative setup used special three-dimensional glass slides to inlay whole ants for analysis—quite a breakthrough considering that previous studies required samples (including ants) to be chopped into thin slices. The additional use of a high-sensitivity mass spectrometer that allows improved accuracy despite changes in sample height allows analysis of complex 3D structures (like that of an ant!).
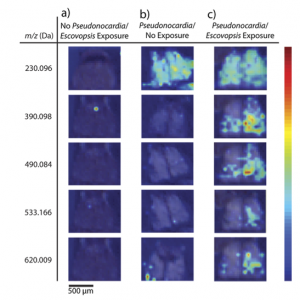
The researchers prepared the ants in environments with their fungal food sources and different exposure to the Pseudonocardia and Escovopsis bacteria prior to mass spectrometry. The ion images they collected (Figure 2) clearly illustrate that the presence of one or both bacteria can drastically alter the variety of compounds produced. One of these molecules was identified as ergothioneine, a chemical with antioxidant properties in vitro that has not been studied extensively in vivo but may play an important role in preventing overaccumulation of the “bad” Escovopsis bacteria.
This novel 3D mass spectrometry imaging technique has opened the door for the discovery of natural compounds in a variety of different symbiotic systems. The authors provide an interesting perspective on the importance of complex environmental interactions for the production of these secondary metabolites. This is especially relevant in a world where the demand for new drug candidates is ever-growing, especially with the rise of antibiotic-resistant bacteria. Perhaps the authors’ technique will be used to shed light on more of these complex microbial communities and identify clinically relevant drugs in the near-future.

