Cover Image: “Shewanella on Hematite”
Photo from Environmental Molecular Sciences Laboratory:
flickr.com/photos/emsl/5788244370
Title: DNA Hybridization To Interface Current-Producing Cells with Electrode Surfaces
Authors: Ariel L. Furst, Matthew J. Smith, Michael C. Lee, and Matthew B. Francis
Year: 2018
Journal: ACS Central Science
DOI: 10.1021/acscentsci.8b00255
As we move deeper into the twenty-first century, the need for clean, “green” energy is ever-increasing. Research into new fuel sources has spanned organisms ranging from plant to microbe in order to fill the increasing energy demands of a technology-driven world. However, while many of these life forms are heavily engineered to pump out a non-native fuel compound, there are others that are naturally suited for useful energy production. Scientists in this article are interested in just such an organism: Shewanella oneidensis.
S. oneidensis is a type of bacteria that can transfer electrons to its environment and generate a functional amount of electrical current — almost like part of a functioning battery! This bacterium has the potential for use in microbial fuel cells due to its ability to convert chemical energy into an electrical output. The one limitation of this process is the requirement for S. oneidensis to attach to a surface by forming a biofilm (a specialized “film” of dense cells), which is time-consuming and difficult to control. Previous research has shown that bacterial growth on electrode plates can be assisted through the use of carbon nanotubes or metal nanoparticles, but biofilm formation is still relatively inconsistent with these methods. As a result, the scientists in this paper decided to turn to a reliable, centuries-old molecule: DNA! DNA strands have a natural affinity for their complementary (perfectly matched) sequence and can therefore be used as a natural “homing beacon” to bring together attached materials (Figure 1).
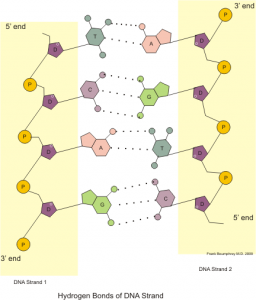
Figure 1: Illustration showing the base pairing between two DNA single strands. These strands have perfect complementarity, allowing researchers to use DNA strand 1 to target DNA strand 2. Image by Boumphreyfr [CC BY-SA 3.0 (https://creativecommons.org/licenses/by-sa/3.0) or GFDL (http://www.gnu.org/copyleft/fdl.html)], from Wikimedia Commons.
The researchers employed a previously-developed technique to prepare live S. oneidensis cells with a coating of single-stranded DNA. These cells were then washed over a gold electrode with a sequence containing DNA with either perfect complementarity or several mismatches (DNA bases that are not perfectly matched). Once the bacteria was affixed to the slide, the cell-electrode complex was tested for current output (Figure 2).
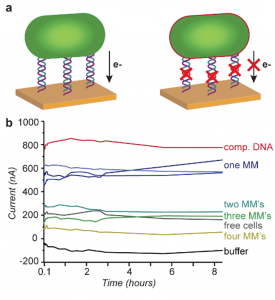
Figure 2: (a) Diagram of bacteria attached to gold electrode through perfect DNA complementarity (comp. DNA, left) or DNA with mismatches (MMs, right). The mismatches reduce electron flow. (b) Current generated at the electrode with different DNA linker sequences. Figure text adapted from paper.
Interestingly, although increasing DNA mismatches (MMs) dampened the current output, this effect was not due to lower levels of bacteria on the electrode. Even with twelve MMs, the DNA had enough complementarity to generate a decent bacterial load (Figure 3). This apparent loss of current was most likely due to disruptions in the DNA helix with each MM. One can think of the DNA as a long electrical wire — although the wire can still connect the electrode and the bacteria, each MM damages the wire slightly until the electrical current is almost nonexistent. A single MM led to a current decrease of 23%, while four MMs led to a whopping 85% dropoff.
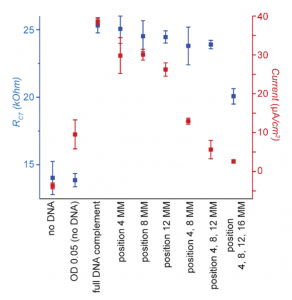
Figure 3: Comparison of relative cell binding (blue, RCT) and current output (red, Current) for different DNA Sequences. Figure text adapted from paper.
The researchers went further to test this DNA “wire” by measuring current before and after cleaving the DNA linkers. They tested enzymes that would cleave all DNA (DNase), cleave only the linker (HinIF), and not cleave the linker (EcoRI). Based upon the results, they found that the DNA linkers were acting like wires, but in a time-dependent manner (Figure 4). Although the enzymes had the expected effect within 1 hour of bacterial incubation on the electrodes, these changes were much less significant at 5 hours and beyond. The researchers speculate that this reduced effect is actually due to the fact that S. oneidensis is capable of forming a biofilm within a 6 hour timespan. This DNA “film” blocks enzymes from accessing the DNA and allows the bacteria to maintain electrical contact with the metal surface.
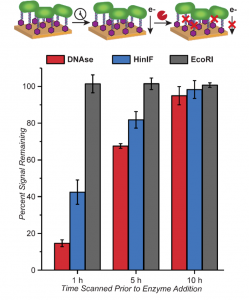
Figure 4: Effects of DNA cleavage on current output. Enzymes for general DNA cleavage (red), specific DNA cleavage (blue), and no DNA cleavage (control, grey) were tested. Figure text adapted from paper.
This article presents a new, innovative method for producing “electroactive” bacterial layers on electrodes. DNA-based targeting has improved the reproducibility of bacterial adhesion while maintaining similar current levels. Hopefully this method can be applied with S. oneidensis and other bacteria for the further development of future microbial fuel cells!