Title: Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme
Authors: Ritu Das, Abhijeet Dhiman, Arti Kapil, Vipul Bansal, Tarun Kumar Sharma
Journal: Analytical and Bioanalytical Chemistry
Year: 2019
https://link.springer.com/article/10.1007/s00216-018-1555-z
What happens when you bring DNA strands, gold nanoparticles, conformation-induced color changes, and a highly-intrusive bacterium together? A field-portable, inexpensive test for one of the world’s greatest bacterial threats, Pseudomonas aeruginosa.
P. aeruginosa is an antibiotic-resistant, opportunistic pathogen that commonly infects patients with weakened immune systems. The World Health Organization (WHO) also considers the microbe’s presence to be an indicator of poor water drinking quality, as P. aeruginosa can infect individuals even at low concentrations, approximately 10-100 colony forming units/milliter. Current diagnosis methods rely on lengthy tests (batch culture), specialized instruments (MALDI-TOF, fluorescent microscopy), and are cost-prohibitive and labor-intensive (ELISA). Enter Das et al.; their proposed replacement method would eliminate these constraints in addition to increasing the speed of diagnosis, portability, cost-effectiveness, and ease of use.
The team, led by THSTI and AptaBharat Innovation, constructed an aptamer-based “NanoZyme” sensor that can target any pathogen. NanoZymes are nanomaterials with enzyme-like characteristics; for example, the gold nanoparticles (GNPs) used in this study have peroxidase-like activity, meaning that the GNPs mimic the horseradish peroxidase enzyme and produce colored products in the presence of peroxide (H2O2). Western blotting and histological staining also take advantage of this color-producing activity.
Das et al. wanted to exploit this activity to detect Pseudomonas. But GNPs always have peroxidase-like activity; any sample applied would cause a color change, regardless if Pseudomonas is present or not. Therefore, the aptamer-designing company did what they do best – designed an aptamer.
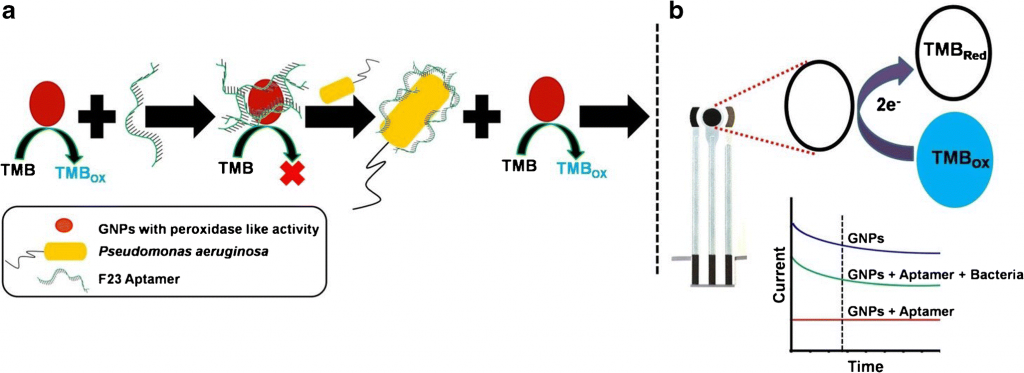
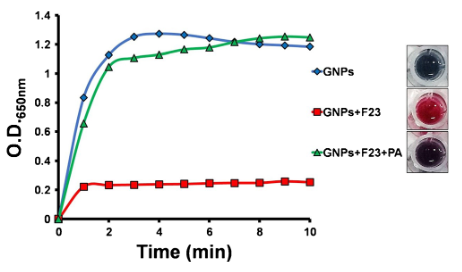
Aptamers are polymers of nucleotides that bind to a specific partner, such as a protein, DNA/RNA sequence, or small molecule. Das et al. designed this single-stranded aptamer, called F23, to change confirmation upon binding to a P. aeruginosa cell. In its original form, F23 adheres electrostatically to the GNP surface and prevents any peroxidase-like activity – producing a vibrant red color (Figure 2). After binding to Pseudomonas, F23 changes shape. The GNPs are then free to oxidize any available substrates, in this case 3,3’,5,5’-tetramethylbenzidine (TMB), producing a deep blue color (Figure 1a; Figure 2). This deep blue color is stable and does not degrade within a 10 minute period (Figure 2).
However, Das et al. did not stop there – they combined their pathogen-specific color test with an electrochemical sensor. TMB is electrochemically active when reduced; by adding sulfuric acid, TMB is converted from its blue, oxidized form back to its non-colored, active form (Figure 1b). The electrical current detected from the active TMB can then be analyzed via a screen-printed carbon electrode (Figure 3).
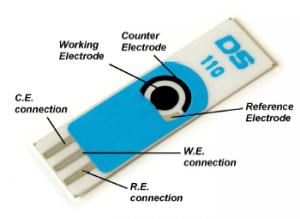
The authors envisioned the protocol as follows: a microliter-sized droplet (containing a mixture of potentially Pseudomonas-contaminated sample, dilute sulfuric acid, aptamers, and GNPs) is placed on the working electrode. A low-voltage electrical current is supplied to the electrode. The amperage difference between the working/counter electrodes compared with the reference electrode is proportional to the amount of reduced TMB generated. Therefore, the greater the amperage generated from the sample, the more Pseudomonas-aptamer interactions there are, increasing confidence in a positive Pseudomonas identification (Figure 4).
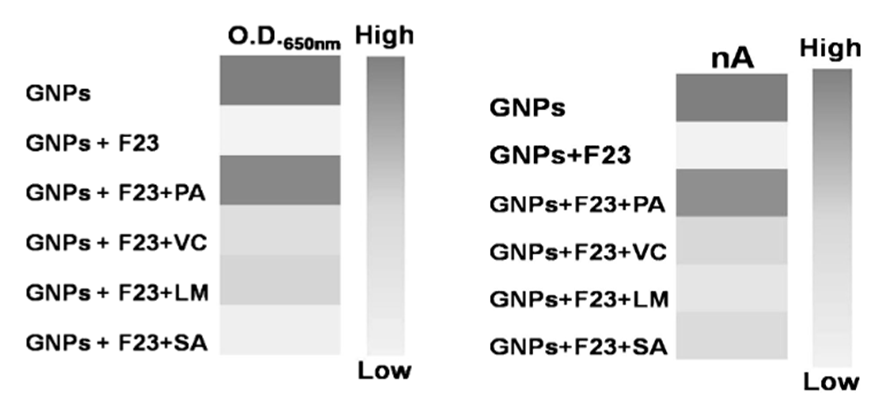
To demonstrate the specificity of their aptamer NanoZyme-based electrochemical sensor, the authors evaluated other common Gram-negative and Gram-positive pathogens. These included Vibrio cholerae, Listeria monocytogenes, and Staphylococcus aureus. The device produced a positive deep blue color change and strong amperage difference only in the presence of P. aeruginosa and GNPs without the aptamer (Fig 4). The strong reaction to GNPs alone affirms that the electrode and color reaction function as normal, but the absence of a reaction when the F23 aptamers are present confirms the validity of the authors’ method. While the linearity of their detection is not ideal (R2 value of 0.93), the authors noted that the lower-end limit of detection was highly relevant to water samples (60 CFUs/mL compared with 100 CFUs/mL), the original goal of the study.Das et al. also showed that compared with currently developed approaches, their system had similar detection limits and took the least amount of time.
Overall, this combination color-electrochemical sensor demonstrates how we can utilize multiple areas of analytical chemistry to address some of today’s most pressing biological issues without expensive or highly technical equipment. This technology may potentially streamline on-site detection for common microbial pathogens in water and food contamination, thereby extending the capabilities of the CDC and the WHO.