Title: Detection, Structure and Function of Modified DNA Bases
Authors: Alexandre Hofer, Zheng J. Liu, and Shankar Balasubramanian
Year: 2019
Journal: Journal of the American Chemical Society
DOI: https://doi.org/10.1021/jacs.9b01915
Featured image and figures Reprinted with permission from J Am, Chem. Soc. Articles ASAP. Copyright 2019 American Chemical Society.
Finding Genes that Fit: Targeting DNA Bases
The launch of the Human Genome Project in 1990 spurred on significant interest in sequencing and editing of the human genome (Figure 1). This project also sparked controversy on how to collect the data required for genome sequencing – though clearly, this data drastically impacted our ability to detect and treat genetic conditions such as cancer.
Figure 1. Example of genomic sequencing represented as the 23 chromosomes.
The 1997 film GATTACA further brought the project into the mainstream spotlight and served to increase ethical concerns on gene sequencing and editing. Much Regardless, scientists cannot deny that this database has allowed us to learn more about ourselves as a species, leading to a wider range of applications, such as ancestry testing and personalized genomic medicine.
While all these are a direct consequence of the sequencing of the primary human genome, other noteworthy features are often not mentioned, such as the chemical changes of DNA. This phenomenon can have an adverse effect on the biomolecule’s structure and function, leading to health issues.
Certain DNA base modifications are well-documented, such as 5-methylcytosine (5mC) Though discovered in 1925, it was not until 1950 that scientists developed a paper chromatography diagnosis to confirm its appearance. Current technology has evolved our diagnostic processes even further; the current standard for detecting and quantifying DNA samples, liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS), can detect the nucleoside modification down to 1 ppm or lower.
In this article, the authors discuss several methods of identifying various changes to nucleobases. Due to the difference in chemical composition of the 4 nucleobases present in the human genome, no universal method exists to detect all changes to the sequence. As such, each type of nucleobase must be treated as a separate sub-class, with their unique processes. This article focuses on the various modified nucleobases and the different methods of detection, utilizing both chemical and antibody-based enrichment methods to improve the detection of such changes. A key issue is the antibody selectivity for specific mutations. The authors solved the limitation by using smaller targeting molecules, which reduce the surface available for non-specific interactions.
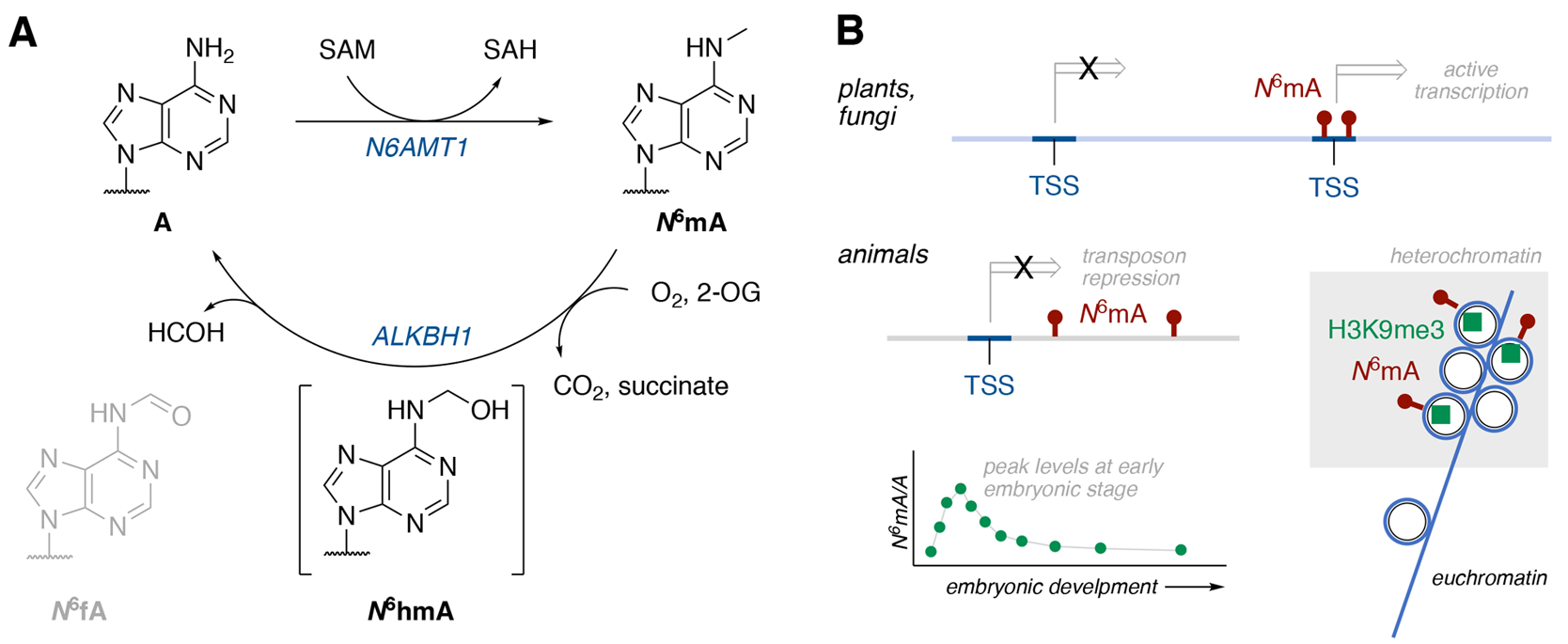
Figure 2. Example of modification of Adenosine (A) and the varying effects in flora and fauna.
Hofer et al. provides a unique insight into the existing nucleoside modifications and their effects, outlining the individual approaches for detection of modified nucleobases. For example, with cystine and adenosine bases, methylation and demethylation seem to be the most common modifications(Figure 2). In these situations, the function of these modified bases has yet to be fully explored. In thymine and guanine, the modifications are more varied. Thymine tends to see the addition on bulky groups at the C5 position. While this is often detected in bacteriophages, it is rarely observed in mammalian cells. Guanine can undergo a variety of modifications, from nitration to oxidation to alkylation. These changes are often a result of DNA damage. The most common modification is the oxidation of guanine, resulting in 8-oxoguanine, which appears to participate in the signaling process and has been observed to have a non-random distribution, indicating that there is something that drives the changes within the genome.
However, many things remain unclear about the genome. includingthe potentially detrimental effect of specific changes in the sequence, Hofer’s, Liu’s and Balasubramanian’s article presents the significant achievements in development of novel methods to detect and map various DNA nucleoside modifications. Yet as science advances and scientists develop novel applications for genomic data, we grew closer to unlocking new tools to detect and treat genetically related health issues.
How Ancestry Testing Works: https://www.washingtonpost.com/videonational/how-dna-testing-for-your-ancestry-works/2016/12/30/9b5ad47a-cea4-11e6-85cd-e66532e35a44_video.html?noredirect=on&utm_term=.fa408a73640c
Trailer, GATTACA: https://youtu.be/BpzVFdDeWyo

