Featured Image Source: https://pixabay.com/illustrations/bacteria-illness-virus-infection-163711/
Primary Source Info
Title: S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate
Authors: Wei Qin, Ke Qin, Yanling Zhang, Wentong Jia, Ying Chen, Bo Cheng, Linghang Peng, Nan Chen, Yuan Liu, Wen Zhou, Yan-Ling Wang, Xing Chen, Chu Wang
Year: 2019
Journal: Nature Chemical Biology
DOI: https://doi.org/10.1038/s41589-019-0323-5
Although traditionally recognized as the body’s infection-defense mechanism, the immune system is now understood to play a complex and fundamental role in all aspects of human health and disease, even in the absence of infection. In a healthy human, immune signals that promote inflammation constantly compete with those that suppress it. Excessive inflammation can harm healthy tissue, causing autoimmune diseases like rheumatoid arthritis or multiple sclerosis (MS), which are increasing in prevalence in the industrialized world1. Excitingly for drug developers, a number of small organic metabolites (compounds found in metabolism) can regulate this immune “posture”. These molecules could be signposts on the road to therapies in several diseases where this immune balance is misregulated.
One such molecule, itaconate, is a metabolic byproduct derived from cis-aconitate (Fig. 1), which is produced in the mitochondria, the “powerhouse of the cell.” This molecule has been known for some time to be anti-inflammatory, hitting the brakes on the body’s immune response. How exactly itaconate produces this anti-inflammatory effect is unclear though.
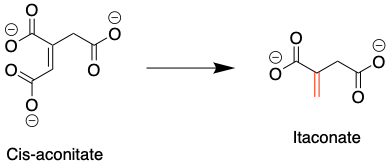
Figure 1: The mitochondrial metabolite cis-aconitate is converted to itaconate. Itaconate’s reactive site is indicated in red. Source: Justin Donnelly
The chemical properties of itaconate offer some clues. Itaconate is an electrophile (meaning electron-loving). In other words, it is electron-poor, and seeks to react with electron-rich molecules. The inside of cells is a highly electron-rich environment, and biomolecules such as proteins have many nucleophiles (nucleus-lovers) for itaconate to potentially react with. The particular structure of itaconate, particularly the double bond highlighted in Fig 1, suggests reactivity with -SH chemical groups, known as thiols, found in proteins on the amino acid cysteine (Fig. 2). The protein KEAP1, known to sense the balance of nucleophilic and electrophilic potential in a cell, was previously shown to react chemically with itaconate in such a manner. This interaction was found to be responsible for some, but not all, of itaconate’s anti-inflammatory effects. But KEAP1 was previously known to be involved in sensing molecules like itaconate. How can we identify other protein partners of itaconate if we do not know where to look for them?
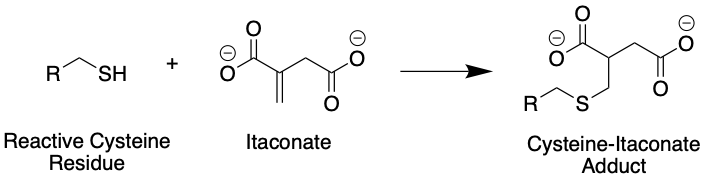
Figure 2: Itaconate can react with cysteines in proteins such as KEAP1, resulting in an itaconate-protein adduct. In this figure, R represents a generic protein backbone. Source: Justin Donnelly
The authors of the Nature Chemical Biology study, Wei Qin and coworkers at Peking University in Beijing, developed a chemoproteomic strategy as a way to scan through thousands of proteins for itaconate-reactive sites in an unbiased way. To accomplish this, itaconate-modified proteins must be selectively enriched from the complete set of a cell’s proteins, known as its proteome, before being digested into smaller pieces and run on a mass spectrometer, which can identify and quantify them.
Enriching modified proteins via chemical methods is difficult, since itaconate is much smaller than the protein it modifies. Well-characterized protein modifications, such as phosphate groups, can be enriched using biological tools like antibodies, but no such tools exist for itaconate. Consequently, instead of enriching itaconate-reactive proteins directly, the authors used a competitive enrichment strategy. By using a synthetic cysteine reactive molecule of their own design, the authors tagged and enriched every reactive cysteine not modified with itaconate. By comparing these enriched proteins to a control group not exposed to itaconate, experimenters can infer whether a cysteine reacted with itaconate, and to what degree, across thousands of proteins at once.
When selecting and developing these probes, the authors began by testing two commonly used cysteine-reactive electrophilic probes, known as iodoacetamide-alkyne and a,b-unsaturated ketone-alkyne (Fig. 3). These probes contain a reactive warhead, iodoacetamide or a,b-unsaturated ketone, that allows them to attach to proteins with nucleophilic cysteines. However, the probes were both too electrophilic to study itaconate modification. At all concentrations, they completely outcompeted itaconate for cysteines across the proteome, meaning no itaconate-reactive cysteines could be identified.

Figure 3: Commonly used cysteine-reactive probes, color-coded according to their components (warhead in red, handle in blue). Source: Justin Donnelly
As a result, the authors sought a less electrophilic probe in order to study itaconate modification and selected an unnatural monosaccharide (sugar molecule) as the scaffold (core chemical structure) for the probe (Fig. 4, left). To fine tune the electrophilicity, the authors tested a peracetylated probe, a sugar molecule in which each hydroxyl (-OH) group was functionalized with an acetyl group (Ac) (Fig. 4, middle). While these probes were too weak to react with a majority of cysteines, liberating a single hydroxyl at the reactive position produced a molecule that modified large number of cysteines in the proteome but could still be competed with itaconate (Fig. 4, right).
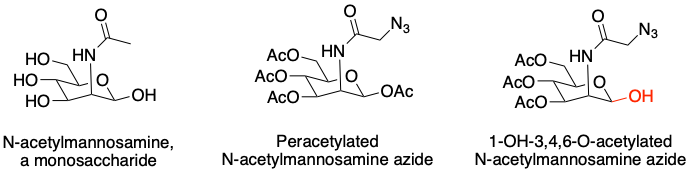
Figure 4: Left: An example monosaccharide. Middle, right: Two probes developed to study itaconate modification of cysteines. The 1-OH at the reactive region of the finalized probe is highlighted in red. Source: Justin Donnelly
These probes also contain an azide (N3) group (Fig. 4) that can be used to attach an enrichment handle using click chemistry, a kind of chemistry specially developed not to interfere with biological components (Fig. 5). This handle contains the chemical group biotin, which is used because it is known to bind very strongly to a protein called streptavidin. By exposing the cells to streptavidin, this property can be exploited to enrich probe-modified proteins.
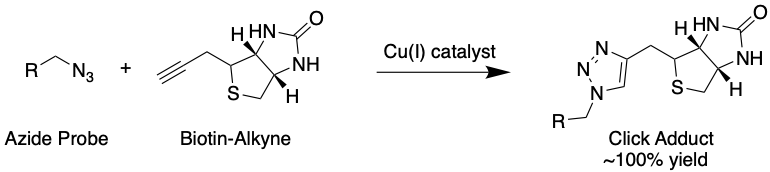
Figure 5: A typical “click” reaction, where an azide and alkyne produce a 5-membered ring adduct catalyzed by copper(I). Click reactions take place under biological conditions with near-perfect yield. In this figure, R represents a probe conjugated to a protein backbone. Source: Justin Donnelly
Using these probes, the authors discovered that, alongside the KEAP1 modifications that had been previously identified, over 260 cysteines in the human proteome were chemically modified by itaconate – a testament to the incredibly complex and varied chemistry taking place in just a single cell. Among the proteins most heavily modified by itaconate in immune cells (other than KEAP1) were enzymes involved in glycolysis, the process by which cells break down the sugar glucose for carbon and energy. As the immune response is energy intensive, and partly facilitated the generation of reactive oxygen species that are harmful to pathogens, limited glycolytic inhibition reduces inflammation.
Targeting glycolysis has had success in autoimmune diseases. The compound dimethyl fumarate, sold as Tecfidera, is a glycolysis-targeting compound effective in the immune disease MS. Tecfidera has an unusually simplistic structure among drugs (Fig. 6), and is a drug version of the metabolite fumarate (also produced in mitochondria) that reduces the excessive inflammation associated with MS by inhibiting glycolysis.
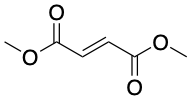
Figure 6: The multiple sclerosis drug dimethyl fumarate, also known as Tecfidera. Source: Justin Donnelly
The results of this study indicate that similar itaconate derivatives are promising candidates to reign in the excessive immune response associated with increasingly frequent immune diseases. Additionally, the probes they developed are not only effective for the study of itaconate, but can be used to explore covalent modification of cysteines by any metabolite with similar electrophilicity.