Title: Beyond Epitope Binning: Directed in Vitro Selection of Complementary Pairs of Binding Proteins
Authors: Eric A. Miller, Ki-Joo Sung, Patthara Kongsuphol, Subha Baniya, Hui Qi Aw-Yong, Vivian Tay, Yuxuan Tan, Farah M. Kabir, Karl Pang-Yeo, Isabel G. Kaspriskie, and Hadley D. Sikes
Journal: ACS Combinatorial Science
Year: 2019
https://pubs.acs.org/doi/10.1021/acscombsci.9b00176?goto=supporting-info
Featured image and figures reproduced or adapted with permission. Further permissions related to this material should be directed to ACS.
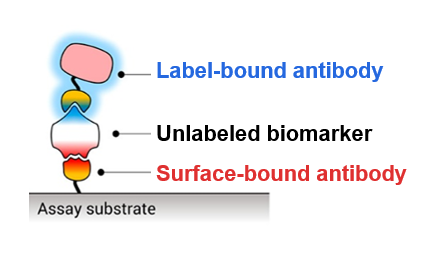
When a person is sick, similar symptoms—fever, chills, exhaustion—could all be signs of many different diseases. Because of this, doctors need tests to determine what disease a patient has so they can administer the proper treatment. Many of these tests rely on the detection of disease-specific biomarkers: in this case, proteins that are present in a patient’s blood, saliva, or urine only if that patient has a specific disease. To detect a specific biomarker, doctors often use antibodies because the original biological purpose of an antibody is to specifically bind a particular region—or epitope—of a protein expressed on the surface of the bacteria or virus causing the disease. One common assay for biomarker detection is called a “sandwich” assay (Fig. 1) and requires two different antibodies that bind different epitopes of the biomarker. One antibody is immobilized by attaching it to a surface, and the blood, saliva, or urine sample is washed over this surface so that the biomarker can bind to the first antibody and stay stuck in place. The second antibody is bound to some sort of label such as a fluorescent molecule which can emit light for detection, and this is washed over the surface to create a “sandwich” where the biomarker is bound between two antibodies. Since the sandwich is still bound to the solid surface, extra fluorescently labeled second antibodies can be washed away without removing the sandwich. Thus, fluorescence is only seen on the surface if the disease biomarker is present, and the use of a second antibody increases the specificity of the assay.
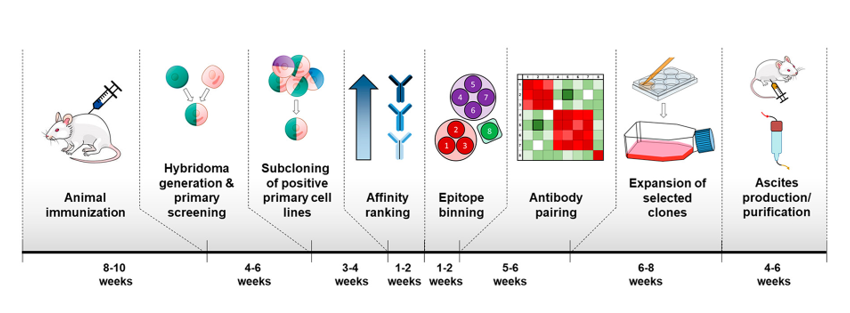
However, at this time, scientists cannot generate completely novel antibodies against new target proteins in the lab. Instead, antibodies must be made in the body of a living animal, such as a mouse or a goat, which can take 8-10 weeks (Fig. 2). In the past, scientists harvested the antibodies directly from the animal (so that a whole mouse or goat would be needed for each batch of antibodies to ever be used), but current technologies allow scientists to fuse the antibody-producing mouse or goat cells with an immortal cell line to create a new cell line called a hybridoma. The hybridoma can be grown in the lab to produce as much of a given antibody as the scientists need for both assay development and subsequent disease detection with the fully-developed assay.
Each sandwich assay requires two different antibodies that bind two distinct epitopes of the same biomarker. Additionally, each antibody also must bind its epitope strongly and specifically, and the two epitopes cannot bind to each other or signal would be seen in the absence of the biomarker, leading to a false positive. It can be extremely difficult and time-intensive to find a successful “affinity pair” of antibodies for one sandwich assay. For example, in a previous attempt which screened 7,680 different hybridomas, only 232 cell lines produced antibodies that were specific enough to the biomarker, and 96% of these antibodies all bound the same biomarker epitope. Because of the difficulty in finding a good affinity pair of antibodies, scientists need a faster, more high-throughput method to screen for good antibodies.
To solve this problem, the current scientists—Miller et al.—developed a quick method to test billions of antibodies against one biomarker to find good affinity pairs for disease detection. This method is called Rapid Affinity Pair via Directed Selection (RAPIDS) process, and the scientists used their RAPIDS method to develop sandwich assays for tuberculosis, Zika virus, and sepsis. Compared to traditional sandwich assay development, which can take 32-44 weeks, RAPIDS only take 10-18 weeks to complete.
RAPIDS begins with a library of yeast cells, where each cell expresses on its cell surface a slightly different antibody against the chosen biomarker. For tuberculosis, for example, the biomarker is called Rv1656, and each antibody tested is a slightly different variant of the rcSso7d antibody that binds Rv1656. Some of these variations bind different epitopes of Rv1656. Some bind tightly while others let go easily, and some bind Rv1656 specifically while others bind many different proteins that may be found in a blood sample and thus aren’t useful for tuberculosis detection. The scientists then combine each yeast cell with the biomarker and sort the biomarker-bound yeast cells from the un-bound yeast cells. This sorting is done by two different methods: Magnetic Bead Sorting (MBS) and Fluorescence Activated Cell Sorting (FACS). In MBS, (Fig. 3A) the biomarker is attached to a magnetic bead before being combined with the antibody-expressing yeast cells. The cells are then flowed through a magnetic machine so that the unbound yeast cells flow quickly passed the magnets while the biomarker-bound yeast cells are held in place. In FACS, the biomarkers themselves are fluorescently labeled, and many of them can bind the antibodies expressed by one yeast cell to create a bright fluorescent signal. These cells are flowed through a machine which can detect the fluorescent light and allows fluorescent cells to go through one channel and non-fluorescent cells through another. Both methods are used to verify that an antibody does in fact bind to the biomarker. Once the cells are separated, scientists can sequence the DNA used to encode for the antibody to know which antibodies are successful.
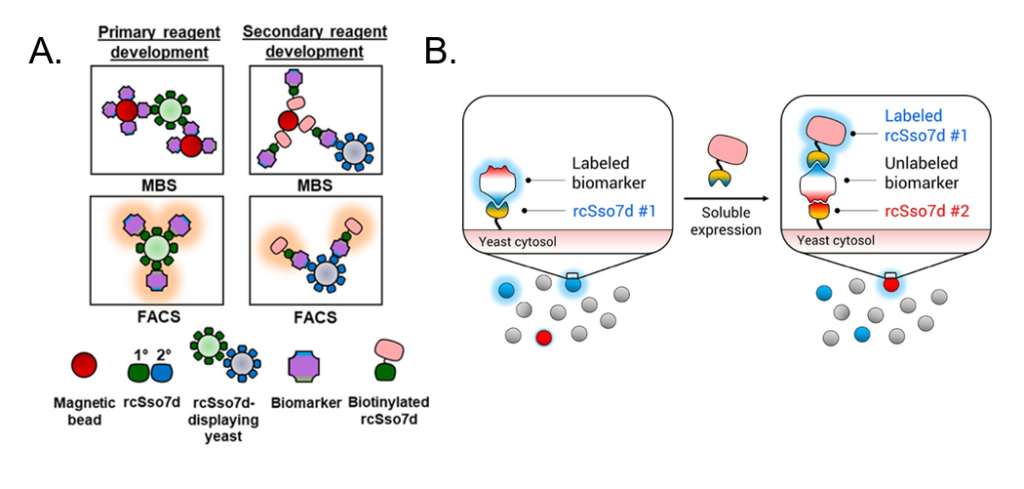
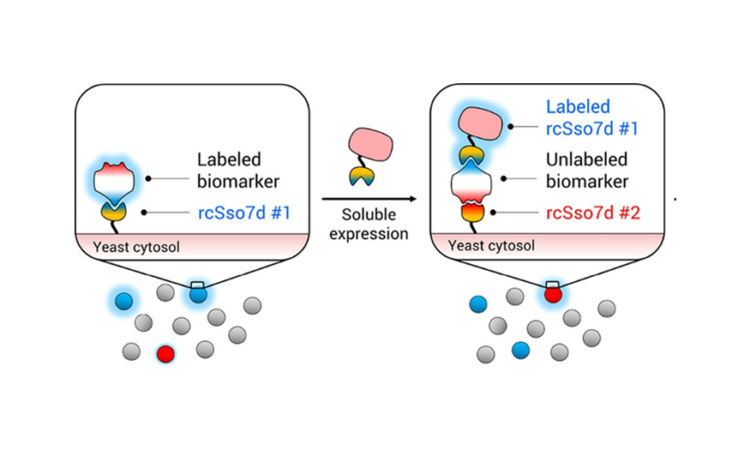
However, a second antibody is still needed to create the sandwich assay. For this portion of the RAPIDS method, the same yeast library is used, but it is combined with a biomarker that is already attached to the first chosen antibody (rcSso7d #1) (Fig. 3B). rcSso7d #1—instead of the biomarker itself—is now attached to the magnetic bead (for MBS) or fluorescent label (for FACS). This setup ensures that the second antibody does not bind to the same epitope as the first, since that spot is blocked by the first antibody that is already bound to the biomarker. In this way, a whole sandwich assay is developed more quickly than was previously possible.
The RAPIDS method is one of many possible strategies currently being developed for selecting antibodies for disease detection. Although it has some limitations—such as relying on the creation of large antibody-expressing yeast libraries—its clear advantage is that it can quickly identify not just one antibody against a biomarker but the entire affinity pair required for a sandwich assay. Miller et al. have already used this method to detect biomarkers for three dangerous diseases, and now scientists will hopefully be able to detect many more for rapid, specific diagnosis and successful treatment.