Article Title: Peptide Ligation at High Dilution via Reductive Diselenide-Selenoester Ligation
Authors: Timothy S. Chisholm, Sameer S. Kulkarni, Khondker R. Hossain, Flemming Cornelius, Ronald J. Clarke, Richard J. Payne
Journal: J. Am. Chem. Soc.
Year: 2019
DOI: 10.1021/jacs.9b12558
Some of the most important biomolecules in our bodies are proteins. They perform a variety of functions including catalyzing chemical reactions, DNA replication, and providing structure, all of which are defined by a specific amino acid sequence. In the body, proteins are synthesized by the ribosome. Over the years, scientists have also created synthetic methods for accessing proteins. This provides an alternative method to obtain, as well as customize, proteins. For instance, these technologies have been used to incorporate unnatural amino acids and post-translational modifications to decorate the proteins in a specific manner. These creations have are important tools for studying cellular biology and also developing drugs.
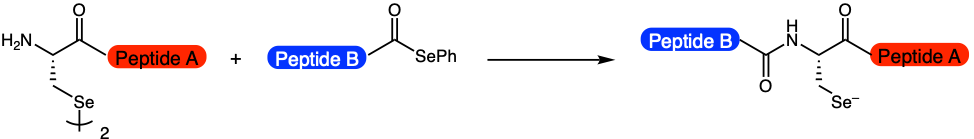
Chisholm et al. have improved upon these already established technologies by developing a method, coined reductive diselenide-selenoester ligation (rDSL), to ligate, or stitch together, two shorter peptides at low nanomolar concentrations (Figure 1). At the site of ligation is a selenocysteine residue. This new tool allows scientists to synthesize lipidated-proteins, or proteins with lipids attached. These proteins have a wide range of applications, including cell signaling. Up until now, this important subset of proteins has been difficult to access, as the peptide fragments are often not very soluble in water and will aggregate if they become too concentrated.
To address these limitations, the researchers optimized multiple reaction parameters (Figure 2). First, they showed that adding pre-reduced phenylselenoate (PhSe) increased the rate of reaction (Figure 2, Step 1). This key discovery was the first sign that the reaction could feasibly occur at low concentrations. Additional optimization led to the development of a one-pot reaction that could generate PhSe in solution while preventing deleterious pathways. This is advantageous, because purification steps are greatly simplified. As a result, the ligation reaction could now proceed to completion at 50 nanomolar concentrations within 96 hours.
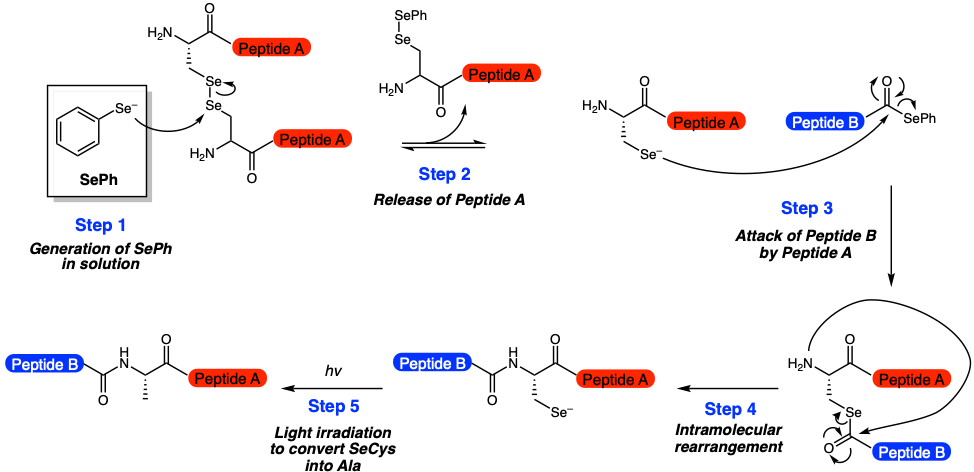
The researchers took it a step even further by exploring methods for deselenization, or a method of converting the selenocysteine residue into alanine. This allows for the original amino acid to be incorporated into the final product. Ultimately, they showed that photochemistry, or illuminating the reaction with 254 nm light for a few minutes, was a viable option (Figure 2, Step 5).
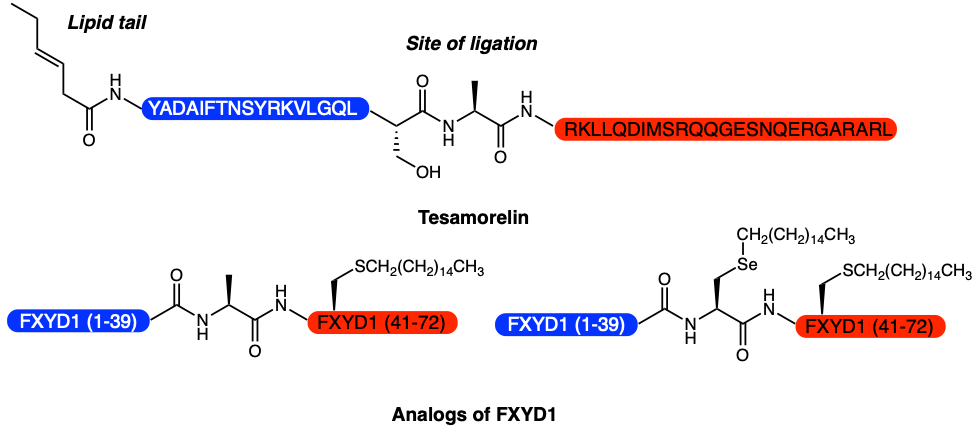
Finally, the researchers were able to apply their newly developed method to synthesize the pharmaceutical Tesamorelin, a lipopeptide that is used to treat HIV-associated lipodystrophy, and analogs of the membrane protein phospholemman FXYD1, which regulates ion pumps (Figure 3). These FXYD1 analogs were later used in downstream experiments to explore how different lipid modifications affect pump activity. This experiment had previously been unachievable as there was no good way of obtaining homogenous samples.
In summary, there is now a validated method for producing lipidated-proteins at nanomolar concentrations in water. Excitingly, this new platform can be used to efficiently synthesize FDA-approved pharmaceuticals. This robust procedure may quicken the rate at which new therapeutics are discovered and screened. In addition, the roles of unknown lipidated-proteins can also be explored, as larger quantities of the protein of interest, as well as analogues, can now be accessed. With these new substrates in hands, additional aspects of cellular biology can now be explored.

