Title: Diversification of protein cage structure using circularly permuted subunits
Authors: Yusuke Azuma, Michael Herger, and Donald Hilvert
Year: 2017
Journal: Journal of the American Chemical Society
https://doi.org/10.1021/jacs.7b10513
Although the environment in an average living cell is drastically different from anything that we observe on a macroscopic scale, one can still find a common parallel with everyday life: package delivery. Certain cells contain proteins that can self-assemble into molecular “cages” to encapsulate materials and shuttle them to various destinations within the cytoplasm. These capsules can also be used to catalyze various chemical reactions within their internal compartment, otherwise known as the lumen. Researchers have adapted these protein containers for a variety of purposes, including biological imaging and delivery of therapeutic agents. However, designing these proteins from scratch can be time-consuming and often produces structures of limited complexity. As a result, the authors of this paper focused on re-engineering existing protein cages to produce “tailored properties” that would be difficult to introduce in a de novo design. The design principles behind these re-engineered proteins could be applied to other protein containers and allow increasingly customizable delivery vehicles for specific applications.
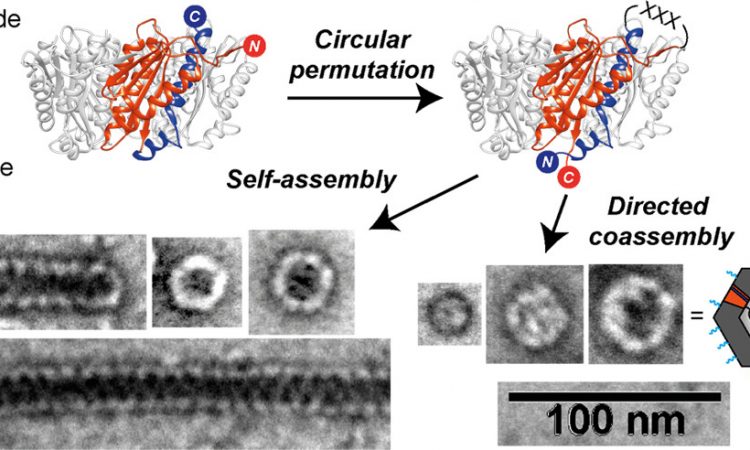
The main principle behind the authors’ design strategy is the concept of circular permutation. In simple terms, circular permutation involves taking a linear sequence and connecting its terminal ends (forming a loop), then cleaving this loop at a distant site to produce a new set of termini. In this paper, the researchers focused on creating circularly permuted mutants of a naturally occurring enzyme, lumazine synthase (AaLS-wt), which can self-assemble into a dodecahedral capsule (or capsid) from 60 identical subunits. Normally, AaLS-wt has its termini on the outside of the subunits, which form the outer surface of the capsid. Through circular permutation, the researchers internalized the termini and created a linker loop on the outside of the protein subunits (Figure 1).
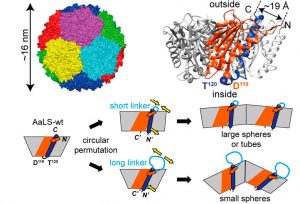
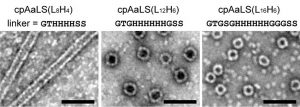
The circularly permuted protein was dubbed cpAaLS and various linker sequences were tested for this design to determine how this region would affect capsid assembly. Using transmission electron microscopy (TEM), the researchers observed that distinct protein structures formed from different linker sequences. A simple change in length was able to shift the structural preference from a tube-shaped capsid to a roughly spherical assembly (Figure 2).
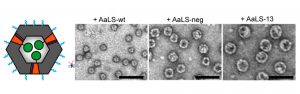
Although the self-assembly of these circularly permuted proteins into roughly spherical structures closely approximates the AaLS-wt shape, the researchers were curious to see what would result from a combination of cpAaLS and AaLS-wt subunits. A hybrid capsid would have distinct advantages because one subunit can easily add fusion proteins to the exterior of the capsid (AaLS-wt) while the other can add distinct fusions to the interior (cpAaLS). After carefully modulating the amount of these two protein subunits in vivo, the authors were able to confirm that fusion proteins formed and were structurally very similar to the original AaLS-wt capsid. Alternate hybrid capsids were also confirmed between cpAaLS and alternative versions of AaLS-wt that have been previously engineered for increased capsid capacity (AaLS-neg and AaLS-13, Figure 3). These structures demonstrate the potential to tune cpAaLS hybrids for different payloads and allow different fusion protein combinations.
These results are promising and suggest that a re-engineering strategy can be used to produce tailored protein capsids for specific applications in synthetic biology. Although much work still needs to be done to verify that these circularly permuted capsids can function similar to the wild-type proteins in vivo, this is an exciting first step towards customizable protein-based encapsulation systems.