Paper Title: A designer enzyme for hydrazone and oxime formation featuring an unnatural catalytic aniline residue
Authors: Ivana Drienovská, Clemens Mayer, Christopher Dulson and Gerard Roelfes
Journal: Nature Chemistry, 2018. https://www.nature.com/articles/s41557-018-0082-z DOI: 10.1038/s41557-018-0082-z
Enzymes are nature’s catalysts, shaped by evolution to perform complex chemical transformations, selectively in our bodies. But they are highly selective about which reactions they catalyse. There just isn’t an enzyme out there for every single reaction chemists want to perform. So many reactions that chemists have created, or want to develop are not known to be enzyme-catalysed. How can we take inspiration from enzymes and use them to make catalysts with new properties and reactivity? In this Chembite, we will look at a recent paper where scientists have managed to do just that.
Enzymes are proteins, made from 20 naturally-occurring building blocks: amino acids. These building blocks contain a range of different reactive functional groups that can be used in catalysis: acids, bases, hydrogen bond donors / acceptors. These reactive groups are located on the side-chains of the amino acids. This could be seen as a limitation, though, because it might be that none of those side-chains can create the right shape/size or chemical environment to allow the substrate to bind and react. As a result, chemists who are designing catalysts are searching for ways of expanding their toolkit.
In nature, enzymes sometimes undergo post-translational modifications in order to get around this problem. Post-translational modifications are reactions that change the side-chains to incorporate new functional groups and reactivity, so that chemists are not limited by the amino acid side-chains available in nature. Another work-around that nature has found is to use co-factors – molecules that are not made of amino acids but are used by the enzyme to assist in catalysis by providing new reactivity.
In this paper, the authors have reported a new way to go beyond the 20 amino acids. They have created an artificial enzyme using an ‘unnatural’ amino acid, i.e. a building block that is not found in nature.
The researchers made their enzyme starting from a protein called a transcription factor. This protein does not behave as a catalyst on its own, its role is to bind to DNA and control the rate that DNA is transcribed into RNA. This protein was selected because it contains a hydrophobic (water-repelling) binding pocket that was identified as being ‘promiscuous’ in binding to a variety of substrates.
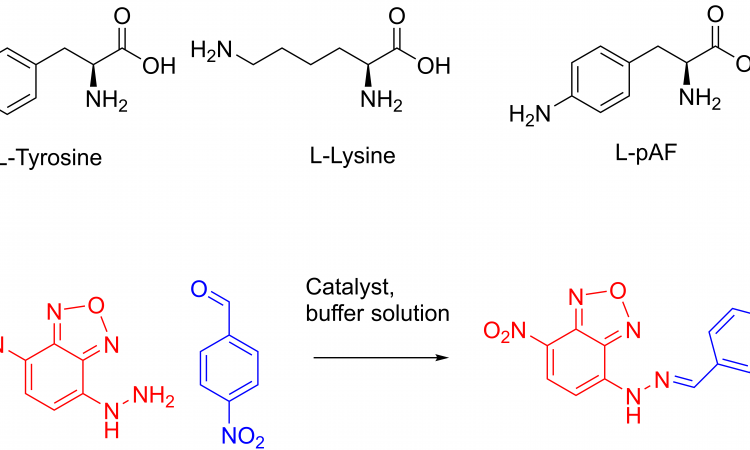
The transcription factor was then modified to incorporate the unnatural amino acid. This was done using expanded genetic code techniques, where the natural genetic code in has been re-programmed to include unnatural amino acids. The unnatural amino acid chosen has an aniline in its side-chain (Figure 1).
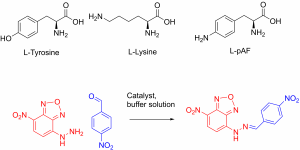
Figure 1: The natural amino acids L-tyrosine and L-lysine next to the unnatural enzyme used in the paper. The pAF abbreviation stands for p-aminophenylaniline.
The aniline side-chain is already a nucleophilic catalyst for the formation of hydrazones (Figure 1). The nucleophile on the amino acid side-chain can attack the electrophilic aldehyde (blue), producing an intermediate that reacts with the hydrazine (red). This particular reaction shown produces a coloured product that absorbs blue light. This allows chemists to monitor the reaction by its colour.
Incorporating the aniline catalyst into the promiscuous binding site of the transcription factor made it a far better catalyst. This is because the transcription factor is able to bind the starting material (substrate) and hold it close to the catalytic site (the aniline). This makes it more likely for the catalysis to take place than if the substrate and catalyst were drifting towards one another in solution.
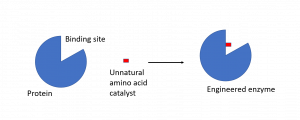
Figure 2: Combining a catalytic amino acid with a protein binding site can enhance catalysis by orders of magnitude.
The efficiency of the catalysis with incorporation of two naturally-occurring amino acids, tyrosine or lysine, (Figure 1) was also tested. These amino acids are also known to behave as nucleophilic catalysts. However, it was shown that the unnatural aniline active site outperformed the natural amino acids.
This work shows how incorporating unnatural amino acids into substrate binding sites can be used to introduce make new enzymes. This opens the door to new reactivities and more efficient catalysts, expanding what nature already has to offer.