Title: Targeting protein-protein interactions for therapeutic discovery via FRET-based high-throughput screening in living cells
Authors: Stroik, Daniel R., Yuen, Samantha L., Janicek, Kevyn A., Schaaf, Tory M., Li, Ji, Ceholski, Delaine K., Hajjar, Roger J., Cornea, Razvan L., Thomas, David D.
Year: 2018
Journal: Scientific Reports
DOI: 10.1038/s41598-018-29685-z
This article is licensed under a Creative Commons Attribution 4.0 International License. Figures and captions were adapted for this post.
Heart failure currently accounts for more than 600,000 deaths each year in the United States, which corresponds to a staggering 25% of all fatalities across the country. Thankfully, advances in medical technology have increased the chances of survival for many Americans who are at risk for heart disease. However, these techniques are mainly targeted at patients who can manage their condition with surgery or lifestyle changes. For those with underlying genetic conditions, heart failure is still a looming threat with few effective treatment options.
In this paper, the researchers focused on a particular protein-protein interaction that has been implicated as a “hallmark” of heart failure. The protein cardiac sarcoplasmic reticulum Ca-ATPase (SERCA2a) is tightly controlled by its inhibitor, phospholamban (PLB) in cardiac tissue. In a healthy heart, PLB carefully regulates the amount of SERCA2a so that the heart muscle can contract (and beat!) at a steady pace. Unfortunately, genetic mutations in PLB can interfere with this well-choreographed interaction, leading to dangerously low levels of SERCA2a and eventual heart failure.
Current treatments for SERCA2a repression frequently focus on increasing the levels of this crucial protein rather than targeting the interaction with PLB. This workaround is not very effective, but was necessary because there were no effective screening methods to select drugs to pinpoint the SERCA2a-PLB interface. The researchers in this paper chose to tackle this problem and have described a FRET-based high throughput screening (HTS) technique to filter out drug candidates that interfere with SERCA2a repression.
The researchers’ method cleverly incorporates a technique called time-resolved FRET (TR-FRET), which is illustrated in Figure 1a. Green Fluorescent Protein (GFP) is attached to SERCA2a and Red Fluorescent Protein (RFP) is attached to PLB. The two fluorophores have a wavelength overlap such that excited GFP can transfer energy to RFP so long as the associated proteins (SERC2a and PLB) are in close proximity. However, instead of looking at the fluorescent output, the researchers looked at the fluorescence lifetime — a very interesting approach! In Figure 1b, one can see that fluorescence of the proteins decreases, or decays, over time. There is a slight difference in these decay waveforms between the bound (GFP-SERCA2a + RFP-PB) and unbound (GFP-SERCA2a only) and this is what the scientists measured when screening drug compounds. This method was chosen because normal FRET was found to be ineffective when used in a similar screen.
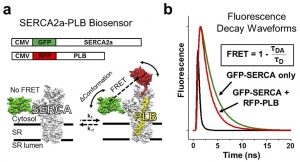
Figure 1: Structure-based high-throughput screening to target the SERC2a-PLB complex. (a) Schematic diagram illustrating the TR-FRET system with both proteins. (b) Graph illustrating the change in fluorescence decay waveforms upon binding of GFP-SERCA2a and RFP-PLB.
After optimizing their screening method, the researchers began testing a library of 1280 common drug compounds. This first experiment pulled out 21 drug “Hits” that displayed a statistically significant change in FRET lifetimes (Figure 2). Digging deeper, the authors looked for false positives by culling out drugs with intrinsic fluorescence or those that targeted the fluorophores instead of the cardiac proteins. The final selection experiment tested for drug activity with SERCA2a and PLB alone (no fluorophores!) to determine if the FRET lifetime changes corresponded with inhibition (decreased levels) of SERCA2a or activation (increased levels) of SERCA2a. This step was essential because the authors could not be sure how changes in fluorescence lifetime corresponded to these levels!
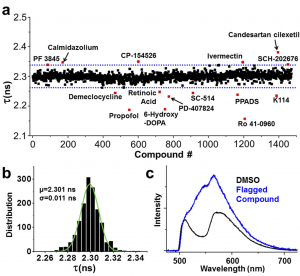
Figure 2: Results of the high-throughput screen. (a) Fluorescence lifetime values for all of the drug compounds screened, with hits highlighted in red. (b) Distribution of fluorescence lifetime values. (c) SERCA-2a-PLB biosensor visible emission spectrum.
After running the protein tests, the authors determined that 2 compounds activated SERCA2a by disrupting interaction with PLB — only a 0.15% true hit rate, but still a success! This result is especially exciting because it demonstrates the power of this new HTS-based technique to identify compounds that can inhibit protein-protein interactions. This method could have far-reaching implications not only in treating heart disease, but treating a number of diseases that were previously thought to be “undruggable.” We look forward to seeing more applications that demonstrate the power of TR-FRET!

