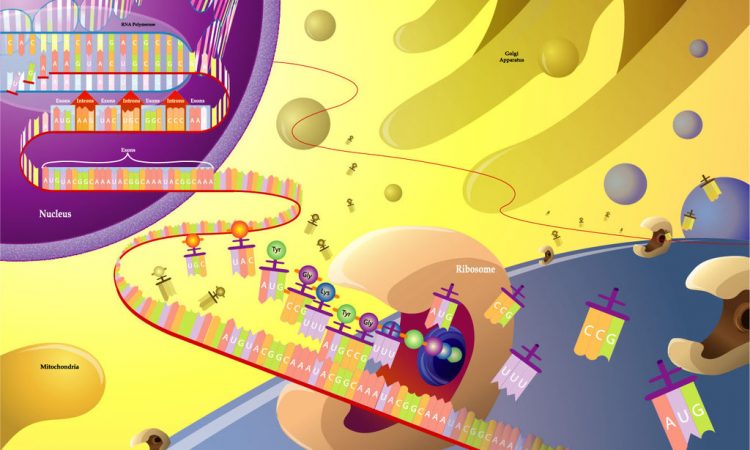
Proteins are macromolecules composed of long amino acid chains folding into complex structures that impart specialized function to the molecule. One of the essential building blocks of living systems, proteins function in nearly every process within the cell—including their own construction!
You may have heard of DNA, or deoxyribonucleic acids. DNA is the double helical nucleotide polymer that makes up your genome. Similarly, RNA is nearly identical to DNA, except that it has one extra –OH group attached to each molecule of the polymer and is typically single stranded due to the added bulk. DNA sits protected within the nucleus of your cells and has to be transcribed to messenger RNA (mRNA) so that can be moved to the cytosol and translated into the proteins that keep the cell running.
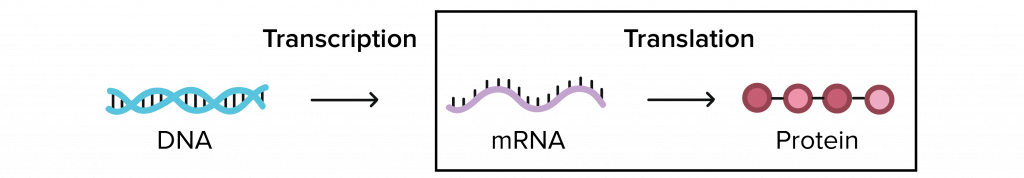
The ribosome is an organelle that makes new protein by translating the nucleic acid code (in three-nucleotide sections called codons) given by mRNA. Composed of two subunits, a large and a small, the ribosome is made of special ribosomal RNA and a variety of small proteins. With the help of metallic molecules called cofactors, these ribosomal RNA and ribosomal proteins fold up and come together to form each unit. The small subunit has to first bind a messenger RNA before attaching to the larger subunit. But then, translation can start when transfer RNA attaches at the first (start) codon. Transfer RNA (tRNA) is what builds the amino acid chain (polypeptide) of the protein. Three-nucleotides long and attached to specific amino acid molecules, tRNA matches the mRNA codon (we call this the anticodon) and transfers the amino acid it carries to the polypeptide chain. From here, the ribosome moves along the mRNA translating one codon at a time. Eventually a stop codon is reached and the new protein is released from the ribosome. New proteins are checked by other organelles for correct folding (because it affects function), but are then able to move around in and between cells to keep your body running. This process of making new protein is critical to keeping you alive!
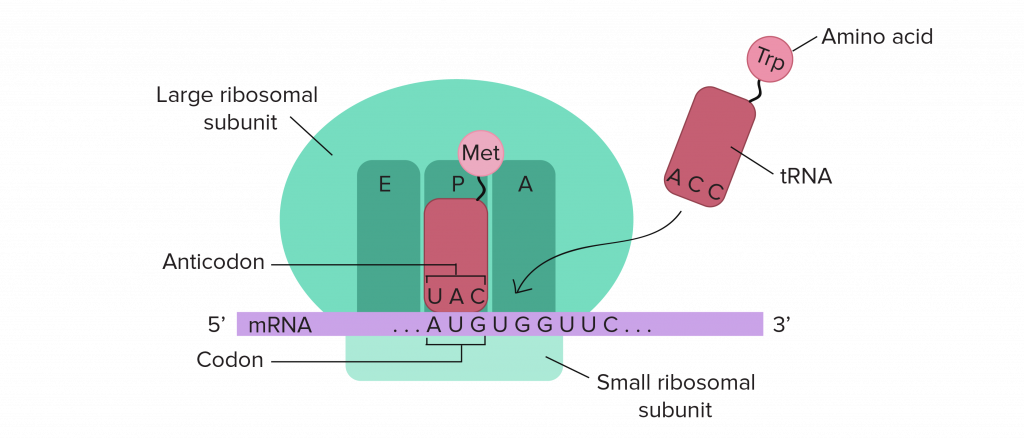
The ribosome and protein synthesis “originated and matured” 4 to 2.5 billion years ago, in the Archean Eon, when the Earth’s crust had just cooled enough for continents to form and life had just barely begun. This early in Earth’s history the atmosphere was anoxic (with very low or zero oxygen level), and metals such as iron and manganese were abundant. Under these conditions, the first photosynthesizing bacteria flourished and gave us the oxygen-containing atmosphere we thrive on today.
The ribosome heavily utilizes magnesium as a metal cofactor in both maintaining structure and mediating translation activity. However, magnesium levels in the Archean atmosphere were much lower than those of iron or manganese. Researchers have already shown that some proteins, such as polymerases, ligases, and ribozymes can functionally substitute Mn2+ or Fe2+ for Mg2+. Moreover, current models suggest that these three metal ions were critical cofactors to ancient nucleic acid function.
Based on this, groups at Georgia Tech hypothesized that under anoxic conditions similar to those of the Archean Eon the ribosome could fold and function with Mn2+ or Fe2+ in place of Mg2+. To test this they first used a method called SHAPE, or selective 2′-hydroxyl acylation analyzed by primer extension. SHAPE uses small electron-seeking chemicals to react with the RNA 2’ hydroxyl group. The overall shape and movement of the RNA affects how flexible, and thus how exposed and reactive, these –OH groups are. By comparing reactivity profiles (SHAPE profiles) researchers can determine the 3D structure of RNA molecules.
In this study, SHAPE profiles after the addition of either Mg2+, Mn2+, or Fe2+ in the absence of O2 all showed folding of the ribosome into nearly identical structures, with the Fe2+ supplemented ribosome coming only marginally closer to the native (Mg2+ ) structure than Mn2+ . So, it’s possible the ancient ribosome was able to fold into a shape highly similar to what we see today.
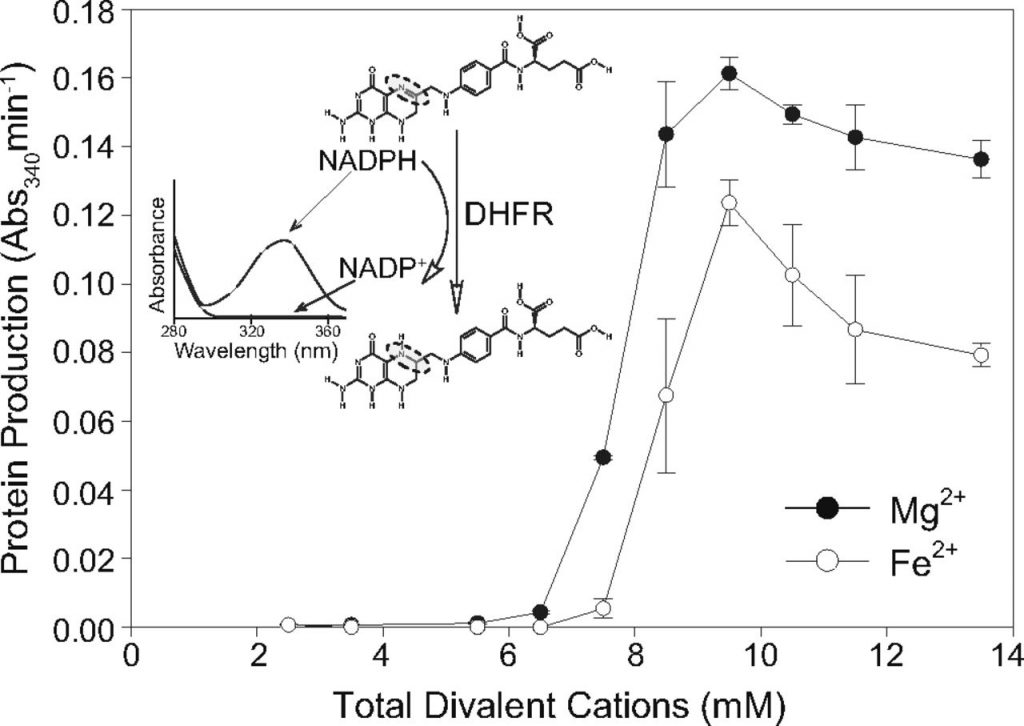
Moving forward to test function, the group used a dihyrdofolate reductase (DHFR) coupled assay to monitor translation. DHFR is a protein that oxidizes NADPH to NADP+. NADPH/+ is a common molecule within cells because it is a reducing agent in synthesis reactions. It is frequently used as a proxy because NADPH absorbs strongly, but the reduced NADP+ does not (Figure 1, inset). So, the relative concentration of NADPH to NADP+ tells researchers how much DHFR has been translated (Figure 1).
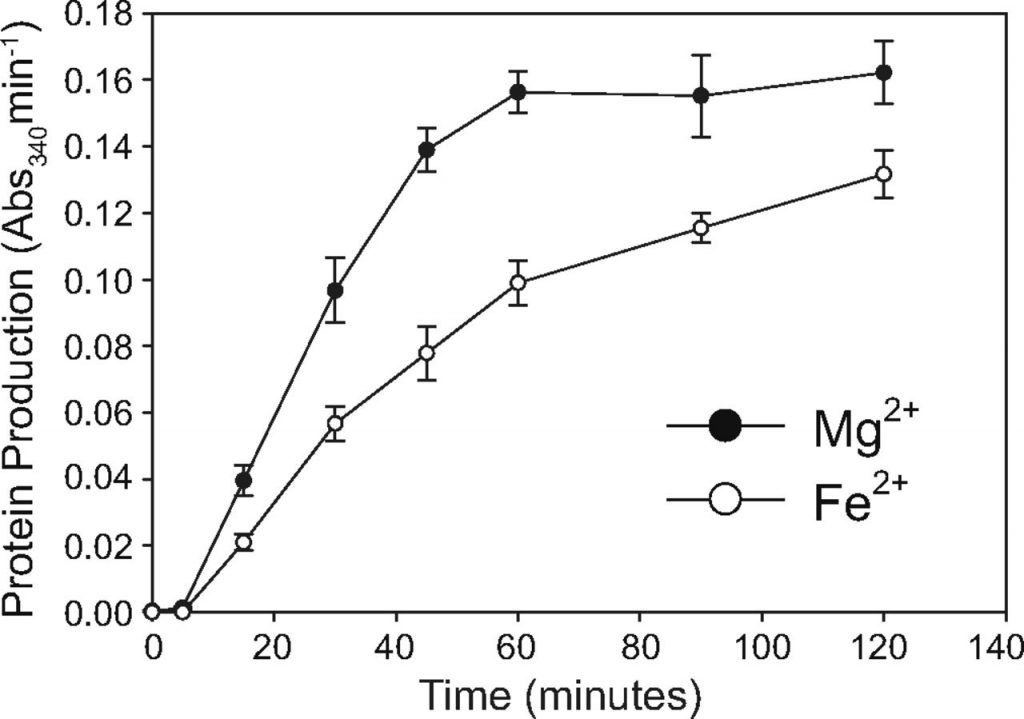
Increasing concentrations of Mg2+, Mn2+, and Fe2+ were added to assembled ribosomes and the progress of translation was monitored. The researchers found that about 7.5 mM of metal was needed for maximum translation activity, and at that concentration the Fe2+ supplemented ribosome was only 80% as active as the Mg2+ supplemented. This was encouraging! To be thorough, other divalent cations were also tested (Co2+, Cu2+, Zn2+), though none but Mn2+ and Fe2+ showed any activity.
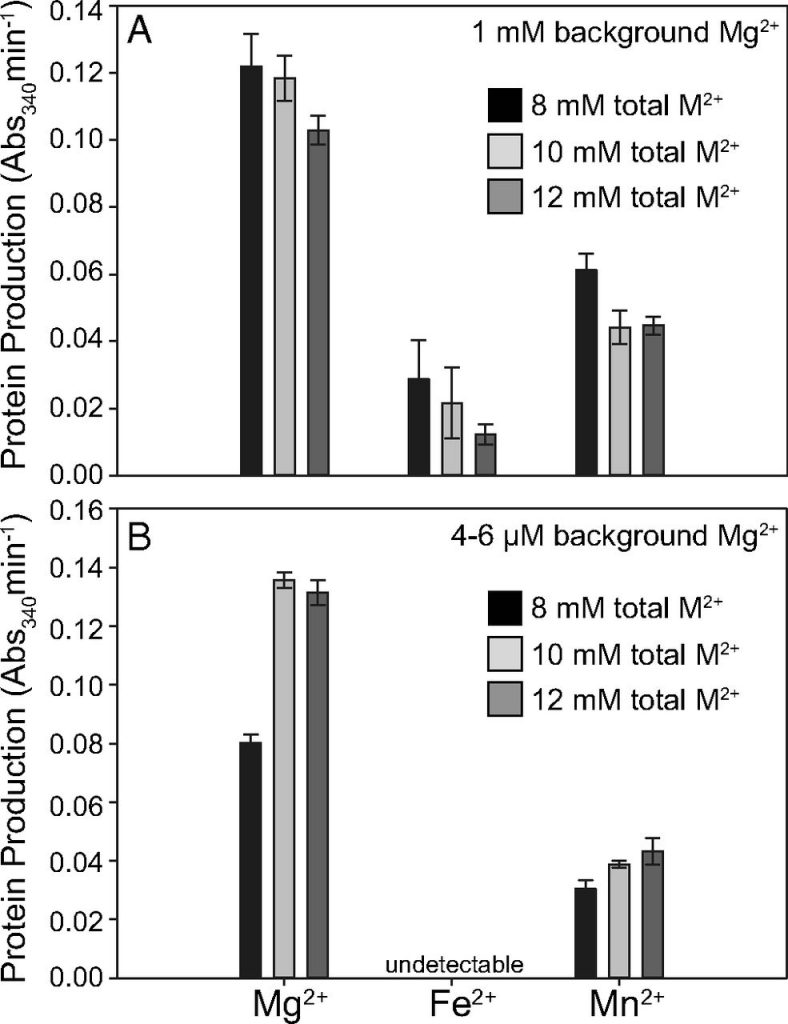
However, to truly mimic Archeon conditions, the background of magnesium used to first assemble the ribosome needed to be significantly reduced. To do this, the researchers washed the assembled ribosomes with divalent cation free-buffer repeatedly until only 4-6uM of Mg2+ remained. While Fe2+ was able to impart activity in a high Mg2+ background, it lost that effect when the Mg2+ was taken away. Fortunately, Mn2+ retained its activity (Figure 3). This was an exciting result because it showed not only that translation can function using a mixture of divalent cations, but that the ribosome could function under ancient conditions.
The translation system– the ribosome and all its metal/protein cofactors– has remained relatively conserved for billions of years. Meanwhile, the bioavailability of divalent cations changes regularly, from longer-term evolution to shorter processes such as oxidative stress. The success of this study highlights the importance of metal cofactors, while raising questions about their evolutionary and physiological role. How is modern biochemistry different from ancient, different from the first biochemical reactions? How has modern biochemistry evolved since life first began? This study gives new insight into these long standing questions. At least in regard to the ribosome, our cells may be performing an ancient protein synthesis reaction.
Title: Multiple prebiotic metals mediate translation
Authors: Marcus S. Bray, Timothy K. Lenz, Jay William Haynes, Jessica C. Bowman, Anton S. Petrov, Amit R. Reddi, Nicholas V. Hud, Loren Dean Williams, and Jennifer B. Glass
Publication Details: PNAS November 27, 2018 115 (48) 12164-12169; published ahead of print November 9, 2018 https://doi.org/10.1073/pnas.1803636115
The original article is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives License 4.0 (CC BY-NC-ND) To view a copy of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/.
Cover Image ©2007-2018 TheElephantMan Creative Commons License via DeviantArt.net