Article Title: Loss of phenotypic inheritance associated with ydcI mutation leads to increased frequency of small, slow persisters in Escherichia coli
Authors: Hingley-Wilson, S. M.; Ma, N.; Hu, Y.; Casey, R. Bramming, A.; Curry, R. J.; Tang, H.L.; Wu, H.; Butler, R. E.; Jacobs Jr, W. R.; Rocco, A.; McFadden, J.
Journal: Proc. Natl. Acad. Sci.
Year: 2020
DOI: 10.1073/pnas.1914741117
In 1942, Hobby et al. discovered that when genetically identical bacteria are exposed to antibiotics, a small population of them continues to survive, or persist. This phenomenon known as bacterial persistence has been observed in nearly all microbes and has immense implications for bacterial resistance to antibiotic therapies. Despite these well-established observations, the mechanism behind persistence is currently unknown, especially because studying these resistant cells, or “persisters”, is difficult.
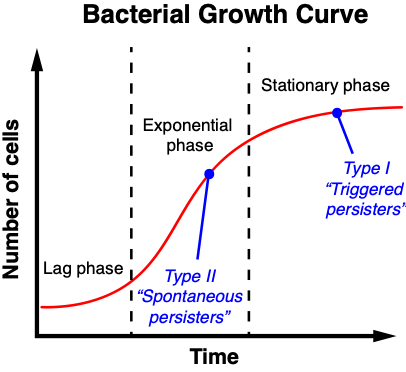
In order to studying this phenomenon, researchers use microfluidics to study and categorize individual bacteria, most commonly Escherichia coli. Past experiments studying E. coli growth showed that most persisters were slow-growing or didn’t grow at all during antibiotic exposure. Ultimately, two types of persisting cells were identified. Type I persisters, or triggered persisters, were generated during stationary phase (phase where there is a constant number of bacteria due to equal growth and death rates). Type II persisters, or spontaneous persisters, were generated during exponential phase (phase during which bacteria are actively dividing) (Figure 1). By studying a mutant strain of E. coli, HipQ, that is capable of generating spontaneous persisters at high rates, Hingley-Wilson et al. begin to unravel just how these bacteria survive.
First, the authors tracked and compared how normal (wild type) and HipQ mutant E. coli grow and divide. By incorporating microfluidics and image tracking of single cells, they found that over multiple generations the two strains did not differ in growth characteristics. More specifically, the division time, elongation rate, size at birth, size at division, and size extensions were approximately the same across the board.
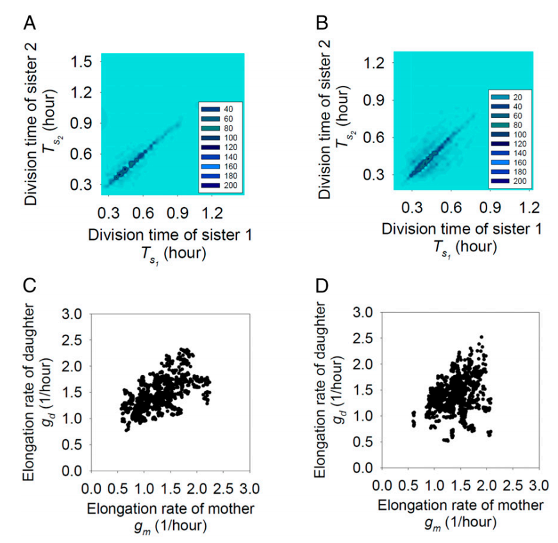
The inheritance of phenotypic variation, or the correlation of observed behavior between mother-daughter or sister-sister pairs, was another story (Figure 2). For wild type E. coli cells, there appeared to be limited correlations in growth characteristics between mother-daughter cells, while sister-sister cells showed similar division times and size extensions. On the other hand, mother-daughter HipQ mutant cells showed loss of correlation of elongation rate and sister-sister pairs showed loss of division time. Altogether, HipQ cells had reduced phenotypic inheritance. As a result, daughter cells are more likely to acquire extreme values of size at birth and elongation rate, enabling them to evade antibiotic death and thus persist.
To take things a step further, they sequenced the genomes of wild type and HipQ E. coli and identified a mutation in the ydcI gene. By introducing this mutation into the wild type strain, they were able to recapitulate the persister phenotype, therefore implicating the ydcI gene in the observation of reduced phenotypic inheritance. Other groups have also associated this gene with the persister trait in other microbes.
Further studies of the relationship between ydcI gene and reduced phenotypic inheritance will be needed. Although the data presented are intriguing, many questions to still remain. For instance, this correlation must also be explored in other disease relevant microbes, such as Mycobacterium tuberculosis, to see if the results can be generalized across the board. Nevertheless, this work sheds light on how the persistence phenotype arises and provides a potential target in future therapeutic endeavors. By understanding how bacteria evade antibiotics, we will be closer to addressing the current antibiotic crisis.

