Article Title: Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels.
Authors: Johnson, E. L.; Heaver, S. L.; Waters, J. L. et al.
Journal: Nat. Commun.
Year: 2020
DOI: 10.1038/s41467-020-16274-w
There is increasingly more press coverage linking our gut microbiome (collection of bacteria and other microorganisms residing in our intestines) to host health and disease. The gut microbiome is one of the densest microbial habitats on Earth and is capable of producing a variety of small molecules. These microbial compounds interface with our own cells, and the constant exchange of metabolites impacts both the host and microbiota. One new link being established is between microbial production of ceramides, a sphingolipid, and host metabolic disorders such as insulin resistance (Figure 1). In this study, Johnson and coworkers provide evidence that Bacteroidetes, one of the most prevalent members of the human gut microbiome, produce sphingolipids that can be transferred to the host intestine and liver.
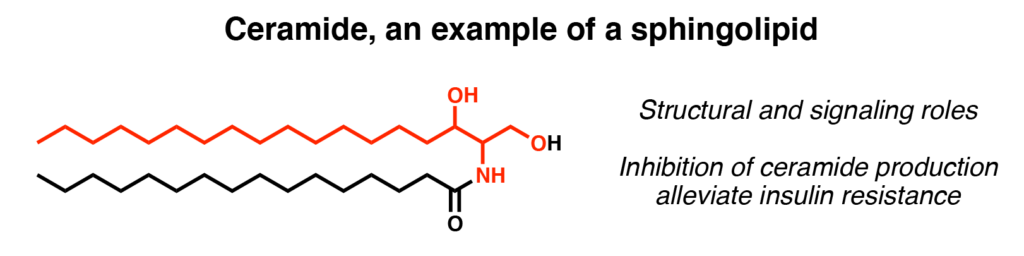
Sphingolipids are a class of lipids with structural and signaling roles in mammals. Humans obtain sphingolipids from the diet or by producing them themselves, in other words, via de novo synthesis (Figure 2A) Certain gut microbiota, including Bacteroidetes, are also capable of producing sphingolipid precursors. These are either the mammalian sphingolipid that contain an even 18-carbon chain (C18) or the shorter bacteria-specific odd 17-carbon chain (C17) (Figure 2B). One major question is whether or not these bacterial-derived sphingolipids also affect host lipid metabolism.
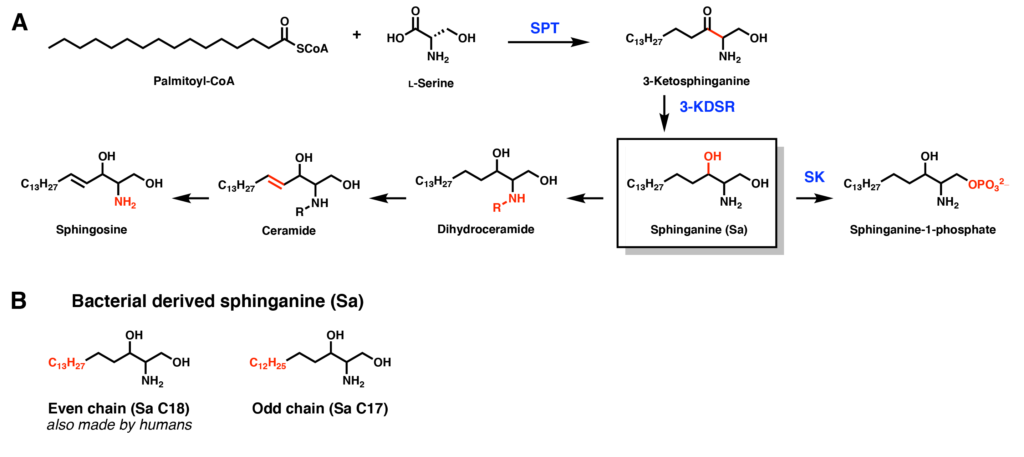
To address this knowledge gap, the researchers took human epithelial Caco-2 cells and dosed them with different amounts of bacterial-derived sphinganine (C17). Using mass spectrometry, they were able to measure how the levels of different sphingolipid metabolites changed in response. If the downstream C18 sphingolipid metabolites (dihydroceramide, ceramide, and sphingosine) decreased in abundance, this would indicate that bacterial sphingolipids inhibit mammalian biosynthesis of sphingolipids. Indeed, these changes were detected, and the researchers noted an overall decrease in de novo C18 sphingolipid production. In addition, the C17 variants of sphingosine and sphinganine-1-phosphate were present, signifying that C17 sphinganine could also be accepted by mammalian sphingosine biosynthesis enzymes.
Although this was an exciting in vitro result, what about in animal models in vivo? The researchers colonized germ free mice (bred mice that have not been exposed to microorganisms) with either wild type (WT) Bacteroides thetaiotaomicron or mutant B. theta with the inactivated enzyme serine palmitoyltransferase (SPT) (Figure 3A). This enzyme is responsible for the first step of de novo sphingolipid synthesis (Figure 2A), so bacteria with this mutant are incapable of producing sphingolipids. With these two strains, effects of bacterial sphingolipid production can be determined.
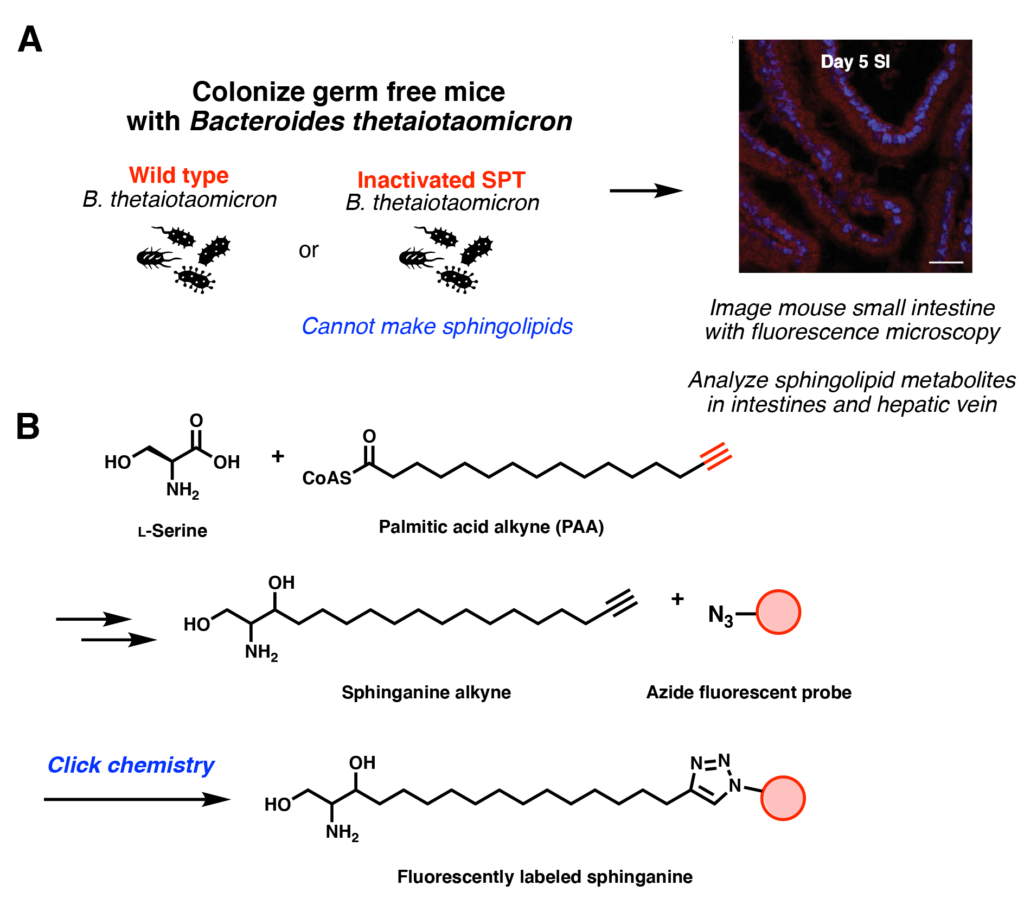
First, however, the researchers had to verify that bacterial sphingolipids are absorbed by the mice’s gastrointestinal tract. They fed the bacteria a substrate analog, palmitic acid alkyne that has a terminal triple bond. This could still be incorporated in the sphingolipid production, resulting in sphinganine alkyne compounds that could be incorporated in mice cells. Through click chemistry, a red fluorescent azide probe could be used to label these metabolites (Figure 3B). Indeed, the small intestinal cells were red after 5 days (Figure 3A-right image), showing that the cell did indeed take up the compound. Additional metabolite sampling from the mice intestinal tract and liver also revealed that sphingolipid production was significantly higher when the mice were colonized with WT B. thetaiotaomicron. This was also the case when an insulin resistant mouse model was used, supporting the idea that bacterial sphingolipid production can impact host metabolism.
As more correlations between the gut microbiome and host are drawn, it is important to understand the mechanisms behind these observations. By understanding how the flow of metabolites between the two parties affects each other, a causative link can be established. These discoveries enable downstream studies to inhibit such processes and alleviate diseases such as insulin resistance.