Title: In Vivo Enzyme Entrapment in a Protein Crystal
Authors: Bradley S. Heater, Zaofeng Yang, Marianne M. Lee, and Michael K. Chan
Journal: Journal of the American Chemical Society
Generally, chemists perform reactions in flasks. A round-bottom flask performs the essential task of holding two or more reagents so they can react in solution. Curiously enough, the concept of a flask works on the nanoscale as well! Materials like metal-organic frameworks (MOFs), zeolites, and aerogels are porous materials that can hold a molecule or set of molecules together in tiny, nanoscopic “reaction chambers” to affect some sort of transformation. Due to the high density of channels and pores that these materials have, a quantity small enough to fit in your hand can have several football fields worth of surface area!
Often when you put reagents in solution and heat them up, many different reactions will occur in addition to the desired one (hence why a series of purification steps are often needed). Carefully designed nanoporous materials can limit these side reactions by acting as scaffolds to hold molecules in specific orientations which encourage one reaction over another. Alternatively, they can act as “bouncers” to limit which molecules can enter the scaffold to access reaction sites within.
By employing these nanoscale reaction chambers, chemists are mimicking the functionality of enzymes in cells. Enzymes contain pockets that fit very specific molecules and can have dangling amino acids that shuffle substrates in one direction and out another. A recent paper by Chan et. al. brings the field of nanoporous materials back to its biological roots by showcasing a system in which one type of enzyme is encapsulated within the porous channels of another much larger crystalline protein.
Because the components are proteins, the assembly of this protein-encapsulated enzyme can be performed entirely in a living bacterium which is reprogrammed to produce both components—doing much of the chemistry for us! The authors used Bacillis thuringiensis, a microorganism that naturally produces a crystalline protein called Cry3Aa. This bacterium is modified to produce a lipase (PML) that comes from the microorganism Proteus mirabilis. Lipases are enzymes that can break down fats like those found in cooking oil. Because the Cry3Aa proteins are crystalline and contain large ~ 5 x 5 nm pores (large by protein standards), they are perfect candidates for a protein scaffold that could encapsulate the much smaller lipase enzyme (Figure 1).
Inside the bacteria, the PML is incorporated into the Cry3Aa as it crystallizes. The size of the PML proteins (6.2 nm x 4.6 nm x 4.5 nm) means that they are small enough to fit snugly inside the pores of Cry3Aa, but encapsulation only occurs when the two proteins are expressed simultaneously by the bacteria. If PML is added to preformed Cry3Aa crystals, PML will not be able to squeeze into the pores.
To confirm that the PML was successfully incorporated into the Cry3Aa crystals, the researchers used fluorescence imaging. Cry3Aa can be visualized with its natural UV fluorescence; PML, which does not emit UV light, can be tracked by labeling it with a dye that emits red light. Thus, when crystals were viewed under the microscope, the presence of fluorescence at each wavelength of light confirmed that both Cry3Aa and PML were present to form 3A[PML] (see Figure 2). Next, they checked for any structural differences between the 3A[PML] and Cry3Aa. X-ray diffraction indicated that inclusion of PML did not influence the structure of the Cry3Aa crystals, implying that the PML molecules were nestled inside the pores of the Cry3Aa. Absence of signal from PML indicated that it had no order within the crystal, an indication that PML molecules might be free to move around in the Cry3Aa pores.
Once the 3A[PML] complex was characterized, researchers tested its catalytic abilities. This showcased the advantages that the assembly has over other protein entrapment methods. Previously, Cry3Aa and PML have been paired together through the direct attachment of PML to Cry3Aa, forming a Cry3Aa-PML fusion catalyst. Compared to Cry3Aa-PML, 3A[PML] can catalyze ten times as much material, likely because the PML is able to be more flexible than when it is adhered to the host protein. On the other hand, compared to the free, unprotected PML protein, 3A[PML] had a three times lower catalytic efficiency –likely due to limited diffusion of substrate to the center of the crystal. However, even with this loss, the trade is a boost in thermal stability and protection from degradation by other enzymes.
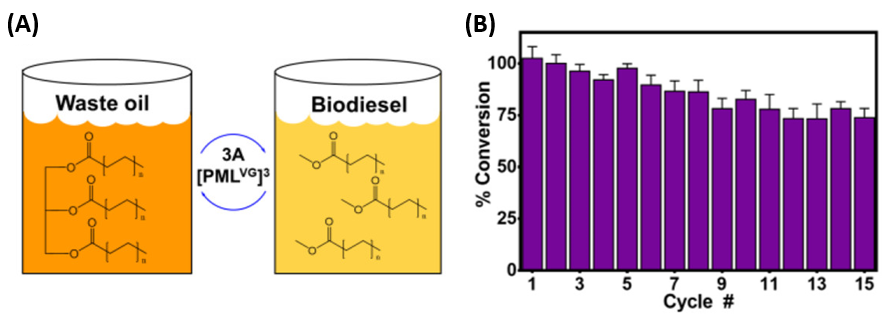
Another advantage of locking a catalyst inside a crystalline framework is added the ability to recycle the catalyst. If a catalyst is just another molecule swimming around in a solution, it may be necessary to use time-consuming separation methods to isolate it from the product it forms. When PML is locked inside Cry3Aa crystals, it is easier to extract the PML from the reaction mixture. The authors demonstrated this by using 3A[PML] to convert used cooking oil into biodiesel (Figure 3). The catalyst was used to convert successive batches of oil into fuel, demonstrating 73% of its original activity upon the 15th cycle.
The design of this nanoporous assembly involved minimal tailoring of the host and guest proteins, as PML is the perfect size to fit inside Cry3Aa pores. Not only that, but researchers already have available a suite of tools that can alter the components involved in this assembly: the genetic material inside a bacterium can be extensively modified to manipulate both the enzymatic activity as well as the shape and size of the proteins. That makes this an exciting method for custom design of catalytic systems comprised of a diverse array of protein-encapsulated proteins.
Featured image and figures reprinted with permission from J. Am. Chem. Soc. 2020, [Online early access]. DOI: 10.1021/jacs.9b13462. Published online: May 14, 2020. Copyright 2020 American Chemical Society.