Title: Cell Membrane-Anchored DNA Nanoinhibitor for Inhibition of Receptor Tyrosine Kinase Signaling Pathways via Steric Hindrance and Lysosome-Induced Protein Degradation
Authors: Jinlu Tang, Cuihua Qi, Xue Bai, Mengmeng Ji, Zhaoting Wang, Yanchao Luo, Shanshan Ni, Tianlu Zhang, Kangdong Liu, and Baoyin Yuan
Publication year: 2023
Journal: ACS Pharmacology and Translational Science
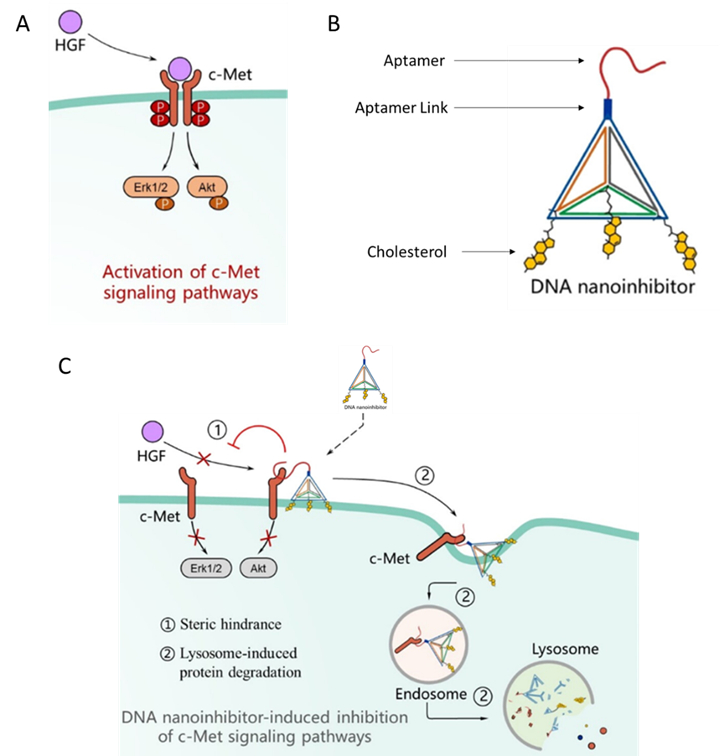
Cells move and multiply through signals received from cell surface proteins called Receptor Tyrosine Kinase (RTK). An example of these proteins is the c-Met protein. When c-Met gets activated by another protein called hepatocyte growth factor (HGF), two c-Met molecules come together, and this initiates a series of signals that causes the cell to migrate and multiply (Fig 1A). In cancer cells, c-Met is overexpressed, and this causes the cancer cells to move to other organs and progress. The current therapies aim to reduce the effect of the enhanced c-Met signaling but these haven’t been effective as cancer cells develop a resistance to these therapies. Tang et al designed a novel therapy that targets the receptor directly using two tools – a DNA tetrahedron and an aptamer.
A DNA tetrahedron is a three-dimensional pyramidal structure formed by DNA strands. It is a stable, rigid nanostructure. The size and structure of the tetrahedron can be controlled by adjusting the length of the DNA sequence. Aptamers are short single stranded DNA or RNA sequences. They can recognize a specific entity and bind to it. As aptamers are made of DNA or RNA molecules, they combine well with the DNA tetrahedron.
To make this construct, they used 4 sequences of DNA which they termed as S1, S2, S3 and S4. c-Met is located on the cell membrane and the membrane is lipophilic. To enhance the binding of the tetrahedron to the cell membrane, they added the lipophilic molecule – cholesterol to sequences S2-S4. These sequences were designed to self-assemble in a tetrahedral geometry. To the tip of this tetrahedral, they added a link through which they connected the aptamer to the tetrahedron. This aptamer was termed as SL1 and it recognized and bound to c-Met specifically. The aptamer and the DNA tetrahedron were connected by a chain called SL1 Link (Fig 1B). The idea behind making this construct is that when it binds to the c-Met receptor, its size would prevent the binding of HGF to c-Met and the dimerization of c-Met. This would then prevent the initiation of downstream signaling which promotes cancer (Fig 1 C).
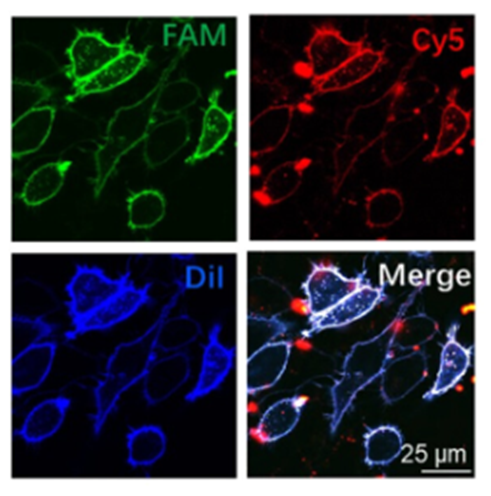
Now that their construct was made, they wanted to see if this construct bound to the cell membrane and c-Met. Further, they also wanted to investigate if binding to c-Met prevented the migration of cancer cells. They tested the construct on liver cancer cells. To confirm the binding of the construct onto the cell membrane, they tagged the cell membrane with a dye called Dil, the S1 chain of the tetrahedron with a dye called FAM and the aptamer link with a dye called Cy5. These dyes are visualized through a microscope and their locations are determined. They found that the positions of where these dyes were located coincided, indicating that the aptamer was linked to the S1 chain and that this construct bound to the cell membrane (Fig 2).
When c-Met is activated by the HGF, a phosphate group gets attached to the c-Met. To measure the inhibition of c-Met by the tetrahedral construct, the researchers added HGF to the cells to activate it and then measured the amount of phosphorylated c-Met in the presence of the tetrahedral construct. They found that at 250nM of the construct, there was no phosphorylated c-Met suggesting that the construct inhibited signaling through this receptor. They also found that in the presence of the DNA tetrahedron, c-Met as a receptor itself got degraded from the cell surface, suggesting that this construct could be used to reduce the levels of the receptor thereby inhibiting further signaling.
A characteristic behavior of cancer cells is migration. Overexpressed c-Met and its activation leads to migration of cancer cells. The researchers showed that the DNA tetrahedron-aptamer construct could inhibit c-Met signaling but did it also translate to inhibiting the migratory behavior? To demonstrate this, they did a couple of experiments. The first being a wound healing assay where a scratch is made on the surface of where the cells grow and the rate at which the cells fill that gap is measured. The other experiment is a transwell assay. Here, the migration of cells across a porous membrane is measured. In both these experiments, the scientists found that in presence of the DNA-tetrahedron, there was less migration as compared to without, suggesting that this construct also inhibited cell behavior.
This research is very exciting in that it has demonstrated how DNA nanostructures are a novel avenue for developing therapeutics. The researchers also showed how this strategy could be generalized to other receptors which makes them easy to implement. Given their ease of constructing, their low cost and the possible customizations makes this an attractive potential therapy option.
Reprinted with permission from ACS Pharmacol. Transl. Sci. 2024, 7, 1, 110–119. Copyright 2023 American Chemical Society.

