Title: DNA-Mediated Peptide Assembly into Protein Mimics
Authors: Fangzhou Zhao, Martin Frandsen, Sabrina Capodaglio, and Hanadi F. Sleiman
Journal: JACS
Year: 2024
J. Am. Chem. Soc. 2024, 146, 3, 1946–1956
https://doi.org/10.1021/jacs.3c08984
Proteins, comprising 20 natural amino acids, play indispensable roles in various life processes. The ongoing computational evolution and improved comprehension of protein folding principles have facilitated the creation of de novo proteins with novel structures and advanced applications. Nevertheless, this endeavor remains a challenge due to the extensive amount of possible sequences and the structural intricacy of proteins. To address the computational challenges associated with larger, more complex systems, using short peptide fragments with predictable structures, including the α-helix, has gained prominence. Coiled-coil motifs are highly stable when two or more α-helices wrap around one another. They are typically composed of seven amino acid heptad repeats and exhibit a hydrophobic core and charged surface residues, forming attractive salt bridges.
DNA, a biomolecule known for its unparalleled programmability and predictability, is a unique scaffold to control assembly due to the predictable Watson−Crick-Franklin base-pairing interactions. The development of these principles has since seen the rapid advancement of DNA nanotechnologies, yielding diverse structural motifs (i.e., tiles, origami-like structures, multiway junctions, and DNA−polymer hybrids), suggestive of our ability to control these materials at the nanoscale precisely. This programmable nature has now been applied to building synthetic proteins to study their folding dynamics and, hence, holds the potential for advancements in biocatalysis.
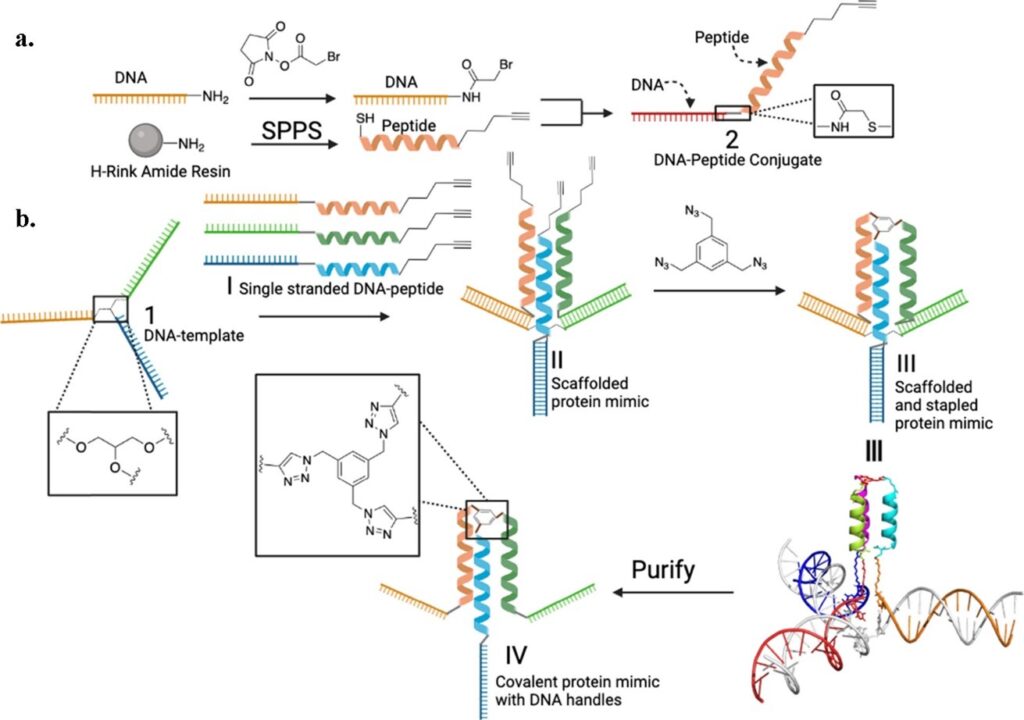
A recent study by Zhao and colleagues introduced a “bottom-up” method to create protein mimics from scratch. In essence, tiny pieces of proteins (i.e., peptide fragments) are connected to synthetic DNA strands using NHS-ester coupling, which can then be carefully arranged into specific shapes using a branched DNA molecular template. The shapes can be predicted by the putative hybridization of these DNA strands to the template. Additionally, the peptide sequences are functionalized with N-terminal alkynes for finally stapling them covalently using a small triazide molecule by click chemistry. The programmed template facilitates the precise assembly of the final protein product in high yield. Additionally, customized DNA strands with varying nucleotide sequences allow for generating different peptide chains and structures, even when the peptide length and interactions alone are insufficient to favor its folding into a tertiary structure thermodynamically. Unlike larger DNA nanostructures, this “trimeric” template ensures the proximity of peptide units, facilitating tertiary structure formation, indicative of our evolving control over the hierarchical structure and our understanding of the nature of the underlying stabilizing interactions (i.e., base-pairing and peptide-peptide interactions).
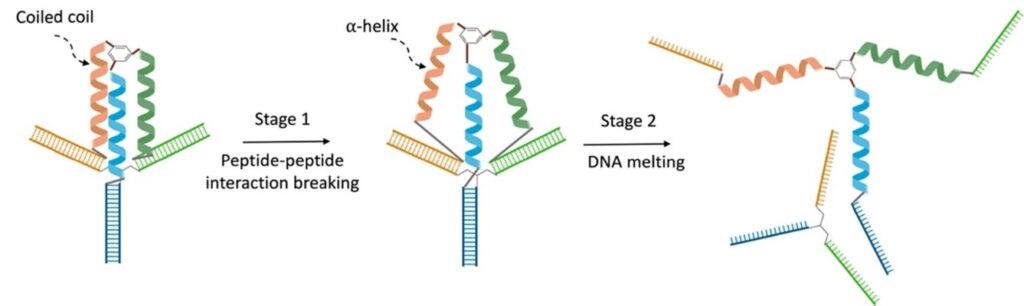
The findings indicated that the stability of these assemblies hinges on two fundamental interactions: DNA base-pairing and peptide-peptide interactions. Thermal stability investigations unveiled a two-step melting process occurring at distinct temperatures (36 °C and 62 °C), suggesting a sequential denaturation of coiled-coil interactions in the peptide sequences and DNA interactions.
This methodology allows us to synthesize an extensive library of protein mimics encompassing different lengths, sequences, and heptads. Notably, this library includes peptides that do not independently form α-helices or coiled-coil motifs. This assembled structure can be denatured to yield fully covalent protein mimics, giving us control over polypeptide conformation. The dynamic tuning of this approach enables the modulation of peptide interactions, transitioning from coiled-coil motifs to random coil peptides, allowing peptides to adopt α-helix or coiled-coil motifs, even with limited lengths and interactions. Think of it like molding clay into different shapes and how you can control the behavior of these assemblies, going from tight coils to more relaxed forms.
In conclusion, the modularity and versatility inherent in this pioneering method mark a promising leap forward in the realm of de novo protein libraries encoded in DNA. This breakthrough not only catalyzes the high-throughput identification of novel therapeutics, enzymes, and antibody mimetics but also facilitates the efficient synthesis of diverse polypeptide architectures. By leveraging different DNA scaffolds as templates, with variations in subunit numbers, peptide orientations, and organic junctions, researchers gain unprecedented access to untapped peptide sequence spaces. The implications of this advancement are profound, promising a new era of exploration and discovery within the intricate landscape of protein design, making it more transformative than before!