Article Title: Electrostatic interactions dictate bile salt hydrolase substrate preference
Authors: Malarney, K. P.; Chang, P. V.
Journal: Biochemistry
Year: 2023
DOI: /10.1021/acs.biochem.3c00210
Our bodies play host to trillions of microbes living inside of our gastrointestinal tract. These microbes are collectively known as the gut microbiome and perform chemical transformations on molecules found within our bodies, impacting our health. For instance, our liver produces bile acids (Fig. 1) that aid in digestion by helping the body absorb lipids and fat-soluble vitamins. These acids are not very soluble in water, so liver enzymes will attach different amino acids to them to increase their solubility. Common amino acids that get attached include glycine and taurine.
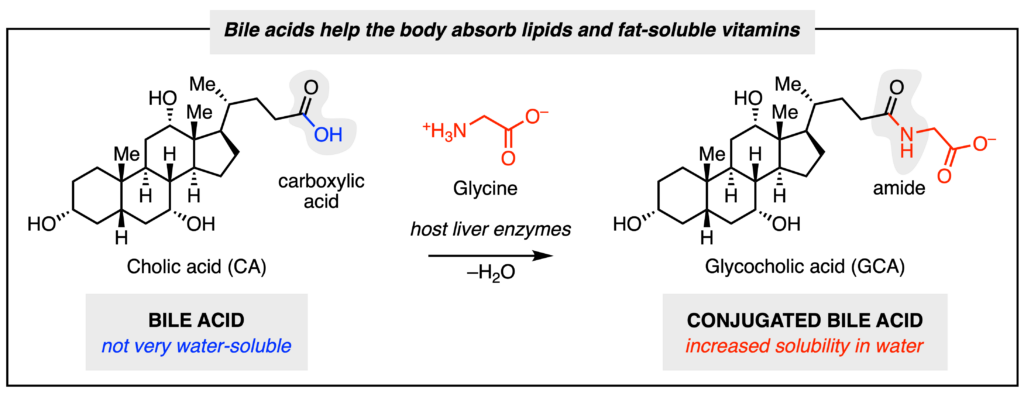
Some gut microbial enzymes are capable of removing, or de-conjugating the amino acid. The enzymes responsible for this chemical transformation are known as bile salt hydrolases (BSHs). Microbes also possess other enzymes to further transform these primary bile acids into secondary bile acids by altering the steroid core (the fused ring section in bile acid molecules). Finally, microbes are also capable of re-conjugating different amino acids back onto the bile acids. These microbially conjugated bile acids (MCBAs) have amino acids such as l‑tyrosine, l‑phenylalanine, and l‑leucine attached (Fig 2). As BSH activity must precede any other bile acid transformations, it is known as the “gatekeeper.” BSHs have different substrate preferences depending on the identity of the steroid core and conjugated amino acid. Given the importance of bile acids, Malarney et al. were interested in investigating how BSHs from two gut microbes, Clostridium perfringens and Lactiplantibacillus plantarum, recognize different MCBAs.
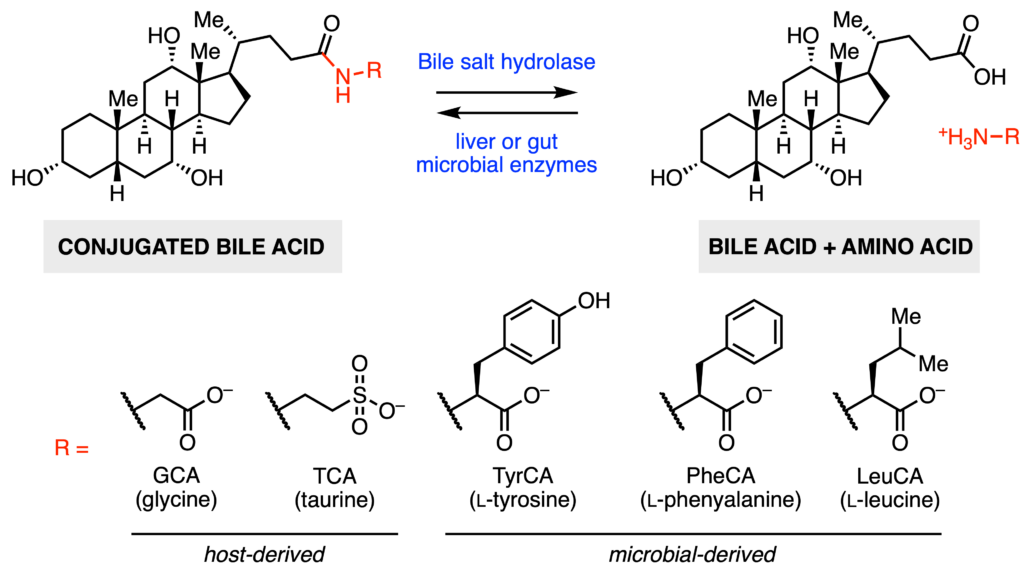
The two BSHs studied were CGH from C. perfringens and BSH1 from L. plantarum. Both enzymes are known to recognize the same steroid core found in cholic acid (Fig. 1). Five different conjugated bile acids were tested for activity: glycocholic acid (GCA – glycine), taurocholic acid (TCA – taurine), and the three MCBAs with either conjugated l‑tyrosine (TyrCA), l‑phenylalanine (PheCA), or l‑leucine (LeuCA) (Fig. 2). To determine which substrates were preferred by the enzymes, the authors measured substrate consumption rates—substrates that disappear faster are preferred. C. perfringens CGH by far preferred GCA and TCA. Of the MCBAs, TyrCA was consumed more quickly than either PheCA or LeuCA. On the other hand, L. plantarum BSH1 had a distinct kinetic profile. Only GCA was rapidly consumed by BSH1. The next two preferred substrates were PheCA and TyrCA. This result was surprising, as this is the first example of a MCBA being more preferred than a host-produced bile acid (in this case TCA).
To better understand the underlying reason for these substrate preferences, the authors used molecular docking to predict which interactions between the enzyme and substrate are important. For C. perfingens CGH, the amino acid portion of the bile acid is exposed to the surrounding polar water. As a result, the charged, polar GCA and TCA molecules are well stabilized (Fig. 3A). Conversely, the MCBAs are quite hydrophobic and do not form such favorable interactions with water. However, since Tyr has a free hydroxyl functional group (–OH), the authors hypothesize that its ability to form a stabilizing hydrogen-bond explains the experimental results.
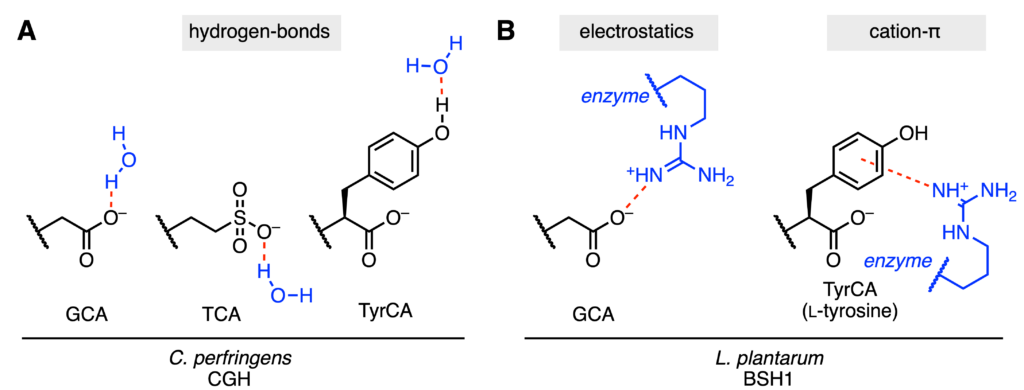
As for L. plantarum BSH1, a critical arginine residue within the enzyme forms important interactions with the bile acid substrate. With GCA and TCA, the arginine can participate in electrostatic interactions (negatively charged GCA/TCA and positively charged arginine) (Fig. 3B). With the MCBAs, cation-π interactions with the aromatic rings of Tyr and Phe can form, which potentially explains the observed substrate preferences. When this arginine was introduced into the protein sequence of C. perfingens CGH, this variant could now process PheCA.
In this article, Malarney and coworkers investigated the biochemical underpinnings of how enzymes can recognize and process different bile acids. The bile acid pool within the human body is diverse, and specific molecules can have different effects on microbes and the immune system. By understanding how these diverse substrates are recognized, scientists can better predict how different gut microbiomes can alter the bile acid pool and impact host health.

