Featured Image Source: https://c7.uihere.com/files/617/602/353/immune-system-infection-regulatory-t-cell-immunology-macrophage-cartoon.jpg
Primary Source Info:
Title: Virus-host interactome and proteomic survey of PMBCs from COVID-19 patients reveal potential virulence factors influencing SARS-CoV-2 pathogenesis
Authors: Jingjiao Li, Mingquan Guo, Xiaoxu Tian, Chengrong Liu, Xin Wang, Xing Yang, Ping Wu, Zixuan Xiao, Yafei Qu, Yue Yin, Joyce Fu, Zhaoqin Zhu, Zhenshan Liu, Chao Peng, Tongyu Zhu, Qiming Liang
Year: 2020
Journal: bioRxiv (preprint)
DOI: https://doi.org/10.1101/2020.03.31.019216
The coronavirus pandemic caused by the virus SARS-CoV-2 has now caused over 100,000 deaths in the United States alone1. However, only a small fraction of people infected develop severe illness leading to death. As a result, researchers want to understand the specific molecular pathology associated with severe COVID-19 in order to more effectively predict at-risk individuals and reduce fatality. Why do some people barely display any symptoms, while others progress to complete respiratory failure?
In order to understand how SARS-CoV-2 infection leads to death in patients with severe COVID-19, it is important to understand how the virus interacts with our immune system. In response to infection, two major arms of immunity become activated: the innate and adaptive immune system. The innate immune system has the ability to sense the presence of infectious agents and respond rapidly through processes like phagocytosis (eating invading or infected cells) and inflammation, which makes the tissue less hospitable to an infectious agent and recruits more immune cells to that site to help fight it. These mechanisms are not specific to any particular pathogen, though, and are often not enough to completely clear it. The adaptive immune system, on the other hand, has the ability to develop highly specific and effective responses to a particular infection, including antibodies, allowing the invader to be swiftly removed with minimal collateral damage to the surrounding tissue. Adaptive immunity can also “remember” pathogens an individual has previously been exposed to, helping prevent reinfection. However, the adaptive immune system requires several days to a week after initial exposure to develop a robust response, so the two halves of the immune system must be able to work together in order to successfully respond to infection (Fig. 1).
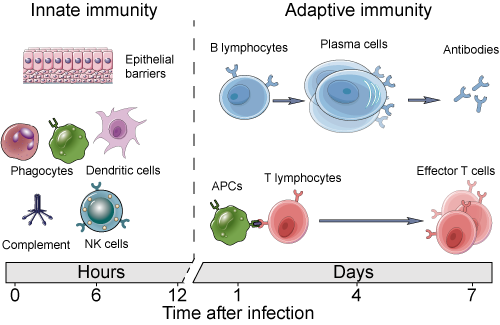
Figure 1: Cell types involved in innate and adaptive immunity, and the time scales on which these systems act to fight infection. Source: https://www.creative-diagnostics.com/images/innate%20and%20adaptive%20immunity2.png
Cells of the immune system use signaling molecules to coordinate their activity in response to infection, known as cytokines. Some cytokines can promote an inflammatory state when released by cells into a tissue, and two of these inflammatory cytokines, interleukin-6 (IL-6) and interleukin-8 (IL-8), are known to play a role in SARS-CoV-2 pathology2. It turns out that the acute respiratory distress syndrome (ARDS) observed in patients with severe COVID-19 isn’t directly caused by the virus, but rather mis-regulation of the immune response. It seems that some patients, such as the elderly, do not produce the same robust, active immune responses as healthier patients. As a result, instead of clearing the virus, a frustrated immune system further increases lung inflammation by increasing levels of IL-6 and IL-8, which recruits more immune cells, leading to more IL-6 and IL-8 secretion. It is this runaway inflammatory process that leads to severe lung damage and respiratory failure. While it is known that COVID-19 patients often experience high levels of IL-6 and IL-8, how exactly SARS-CoV-2 infection leads to these increases is still unclear.
Many biological processes, including those that power the immune system, make use of protein-protein interactions (PPIs). Molecules in liquid and solid phases are held together by physical attractive forces, which are weaker and shorter-lived than chemical bonds and are major determinants of the properties of a substance. These interactions are also responsible for protein folding and PPIs. Chemical technologies are incredibly powerful tools to observe these interactions, and the development of mass spectrometry-based proteomics has therefore greatly improved our ability to identify and characterize new PPIs, Li and coauthors at Shanghai Children’s Hospital decided to study the interactions between SARS-CoV-2 proteins and host (human) proteins to see if they could better understand how SARS-CoV-2 causes runaway inflammation and ARDS.
The most straightforward way to identify the PPIs a particular protein makes is to allow it to form interactions with proteins in a complex mixture, such as the proteome of a human cell, and then extract the resulting multi-protein complexes by enriching, or “pulling down”, the protein of interest. While these methods can identify all interactions a given protein is capable of making, it does not necessarily report on their biological relevance, so it is important to validate any PPIs discovered by this method to confirm their biological function. Nevertheless, these methods are a powerful strategy to identify new molecular biological mechanisms with limited prior knowledge. The authors applied this strategy to SARS-CoV-2 by attaching a tag, known as 3xFLAG, to each of the viral proteins, which facilitates their enrichment. They then expressed each of these proteins in a human cell, which allowed them to pull out protein complexes held together by PPIs between viral and human proteins. The proteins in these complexes were then identified by mass spectrometry (Fig. 2).
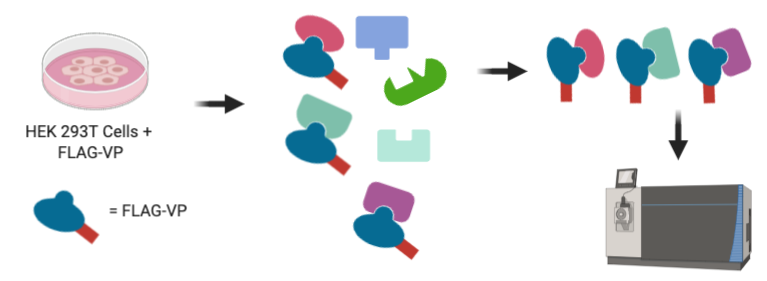
Figure 2: Human HEK 293T cells are programmed to express each SARS-CoV-2 viral protein (VP) with an attached FLAG tag. After cell lysis, FLAG-tagged viral proteins are enriched along with their interaction partners. These interaction partners are then identified using a mass spectrometer.
Some of the most interesting interactions the authors identified were between two viral proteins, known as nsp9 and nsp10, and the human protein NKRF. NKRF is a transcription factor that helps cells regulate the amount of a gene that is produced, or expressed, in a cell. Specifically, NKRF represses genes that are stimulated by another transcription factor NF-kB, which is activated during inflammation. Since levels of both IL-6 and IL-8 expression are increased by NF-kB, the authors investigated whether the interaction of nsp9 and nsp10 with NKRF played a role in their increase in COVID-19 patients. The authors expressed either nsp9 or nsp10 in A549 cells, which are derived from lung tissue. They then analyzed the levels of IL-6 and IL-8 that those cells released into their media. Both nsp9 and nsp10 expression resulted in elevated levels of both IL-6 and IL-8 compared to untreated cells, while expression of nsp12, nsp13, or nsp15 (other SARS-CoV-2 proteins) had no effect (Fig. 3). This confirmed the authors hypothesis that nsp9 and nsp10 played a role in IL-6 and IL-8 increase in severe COVID-19, and this increase is probably related to its interaction with NKRF.
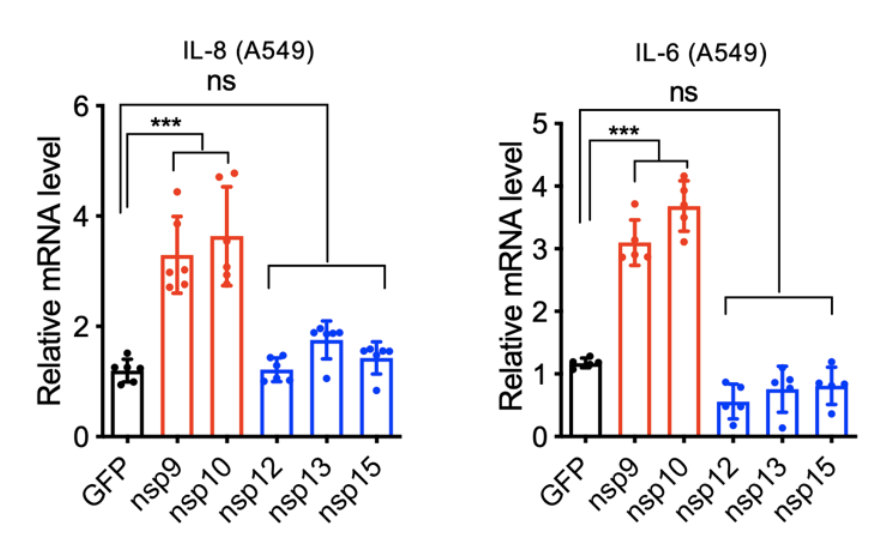
Figure 3: Relative levels of mRNA encoding either IL-8 (left) or IL-6 (right) in A549 lung cells engineered to express different SARS-CoV-2 viral proteins.
Finally, the authors looked at the levels of IL-6, IL-8, and other proteins in immune cells collected from patients with mild or severe COVID-19. The immune cells most commonly isolated from patients are known as peripheral blood mononuclear cells (PBMCs), which can be enriched from whole blood. These cells include T and B cells, responsible for the adaptive immune response, monocytes, which are the precursor cells to phagocytes of the innate immune system, and several other immune cell types. The authors observed elevated levels of IL-6 and IL-8 in PBMCs from patients with severe COVID-19, but not patients with mild disease, consistent with previous results correlating IL-6 and IL-8 to COVID-19 severity (Fig. 4). They also observed elevated levels of proteins responsible for the release of and response to these inflammatory cytokines and depressed levels of T cell proteins, which is likely related to the weakened adaptive response observed in patients with severe COVID-19. Altogether, these data point to an important role of certain SARS-CoV-2 proteins in the progression of coronavirus infection to ARDS and death, and identify potential new drug targets specifically aimed at preventing virally-induced respiratory failure.
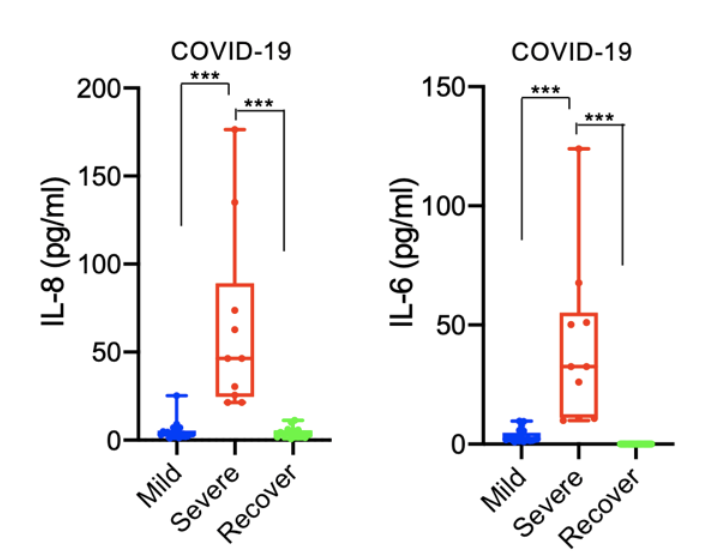
Figure 4: Concentration of IL-8 or IL-6 in PBMCs from patients with mild COVID-19, severe COVID-19, or who have recovered from COVID-19.
Other References:
- The New York Times. (2020, June 3). Coronavirus in the U.S.: Latest Map and Case Count. https://www.nytimes.com/interactive/2020/us/coronavirus-us-cases.html#hotspots
- Conti, P., Ronconi, G., Caraffa, A., Gallenga, C., Ross, R., Frydas, I., & Kritas, S. (2020). Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. Journal of Biological Regulators and Homeostatic Agents, 34(2). https://doi.org/10.23812/CONTI-E

