Primary Source Info:
Title: SARS-CoV-2 Proteome Microarray for Mapping COVID-19 Antibody Interactions at Amino Acid Resolution
Authors: Hongye Wang, Xin Hou, Xian Wu, Te Liang, Xiaomei Zhang, Dan Wang, Jiayu Dai, Hu Duan, Shubin Guo, Yongzhe Li, Xiaobo Yu
Year: 2020
Journal: bioRxiv (preprint)
DOI: https://doi.org/10.1101/2020.03.26.994756
The worldwide death toll from the pandemic disease COVID-19 has now surpassed half a million people, sparking a global interdisciplinary scientific response. One of the most important goals of that response is vaccination. When we become infected, such as by a virus or bacterium, our immune system recognizes the infectious agent as foreign and initiates a response. In the course of this response, a component of the immune system known as adaptive immunity develops highly specific responses to the invader, allowing it to rapidly recognize and clear it with minimal collateral damage to the host. Vaccination involves “training” the immune system by exposing it to molecular signatures associated with a particular pathogen but delivered in a non-infectious form. Thus, if the vaccinated host is ever exposed to a pathogen, the immune system will already be fully prepared, and the infection will fail to take hold. Broad vaccination is fundamental to modern life: as the COVID-19 pandemic has demonstrated, the lack of an effective vaccine against even one highly transmissible disease can have devastating effects on globalized society.
Vaccination can be accomplished in different ways. Early vaccines used attenuated versions of a disease-causing agent, although these strategies carry the greatest risk of actually causing the illness in the host, especially if their immune system is compromised in some way. However, modern advances in immunology enabled by chemical technologies have led to a significantly improved understanding of the molecular basis of pathogen recognition by the adaptive immune system. This has enabled the design of more advanced vaccines composed of only the necessary molecular signatures for pathogen recognition, which are both safer and more effective than attenuated strains (figure).
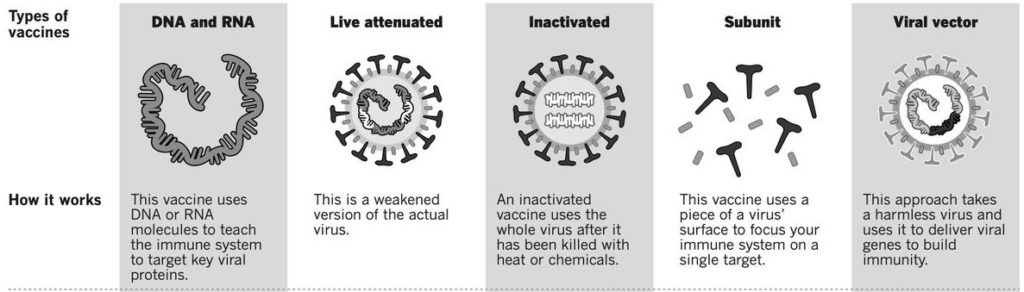
Figure 1: The various classes of viral vaccines. Different forms of whole virus, subunits to elicit specific responses, or the DNA or RNA encoding those subunits, can all be used.
Image source: https://pbs.twimg.com/media/EZ6tyiOVAAAvjPX.jpg
One of the most important components of the adaptive immune response is the antibody response. Antibodies are proteins that have the ability to bind very strongly and very selectively to a particular target. A functional immune system’s B cells will raise antibodies against nearly any foreign substance it encounters, and once it has, clones of these B cells will return to the bone marrow as memory cells, where they can live for decades. Cells like these are the basis of long-term immunological memory. However, not all antibodies are created equal: a single protein, let alone an entire virus or bacterium, can result in antibodies targeting many different molecular signatures, known as epitopes. The particular epitope an antibody interacts with has significant implications for its functionality. The most desirable type of epitope is known as a neutralizing epitope. An antibody targeting a neutralizing epitope binds a region on its target molecule that is so disruptive to its function that the infectious agent becomes unable to infect the host, resulting in immunity. Additionally, the immune system is known to preferentially raise antibodies against certain “immunodominant” epitopes, while showing difficulty raising antibodies against others. Understanding which epitopes on a virus like SARS-CoV-2 (the causative agent of COVID-19) are both neutralizing and immunodominant is critical to the selection of the specific molecular signatures to use in a vaccine.
To address the question of which SARS-CoV-2 epitopes are immunodominant, Hongye Wang and coauthors developed a SARS-CoV-2 peptide array which can be used to analyze antibodies from patients with COVID-19. To make this array, the authors chemically synthesized 15 amino acid peptides corresponding to the entire SARS-CoV-2 proteome, with 5 amino acid overlaps between individual peptides. These peptides were then conjugated to biotin, which is a molecule that interacts very strongly with the bacterial protein streptavidin. Peptides are then “printed” on a streptavidin-coated plate by dropping small droplets onto specific points of the plate (figure). The authors also included two full-length SARS-CoV-2 proteins as well as 5 truncated ones, all expressed in cells (instead of chemically synthesized), as controls to ensure the antibody interactions were properly captured by the chemically synthesized peptides.
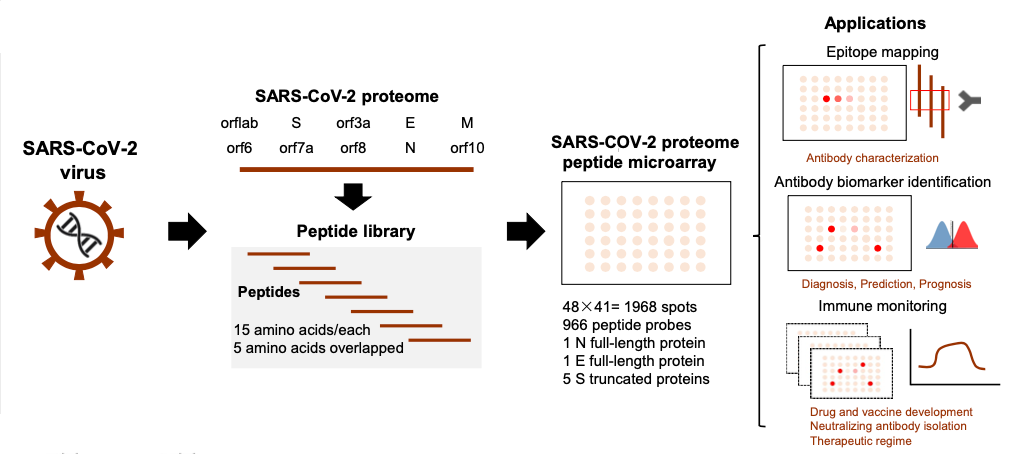
Figure 2: Overlapping peptides covering the SARS-CoV-2 proteome are “printed” at specific points on a glass slide. The slide can then be treated with SARS-CoV-2 antibodies to analyze their binding characteristics.
To validate the array, the authors screened serum antibodies from 10 COVID-19 positive patients by treating the array with the antibodies. A fluorescent antibody targeting the constant region of the serum antibodies, an epitope which will be found on all human antibodies regardless of what they target, can be used to visualize which peptides the patients’ antibodies are binding to on a microscope, and thus determine their specificity. This makes the SARS-CoV-2 peptide array the first proteomic method to profile the global epitope preferences of the COVID-19 antibody response in an unbiased way.
From their initial experiments with these patient samples, the authors identified multiple immunodominant epitopes associated with several SARS-CoV-2 proteins, including the spike protein that facilitates viral cell-entry. Additionally, one of the immunodominant epitopes they discovered on the spike protein was known to be an important component in the protein’s receptor binding domain, the domain that engages the host receptor ACE2 leading to viral entry. They also found that the two major classes of antibodies raised against SARS-CoV-2, IgM and IgG, displayed different epitope preferences (figure). IgM antibodies are usually raised against a virus early in infection, but “convert” to IgG once the antibodies bind more tightly and specifically. Both of these findings will aid vaccine developers in the selection of vaccine epitopes that effectively induce a neutralizing antibody response. Additionally, the technology will enable larger studies into the SARS-CoV-2 antibody response, which could help us better understand the disease progression as well as the broad range of COVID-19 severity across different patients.
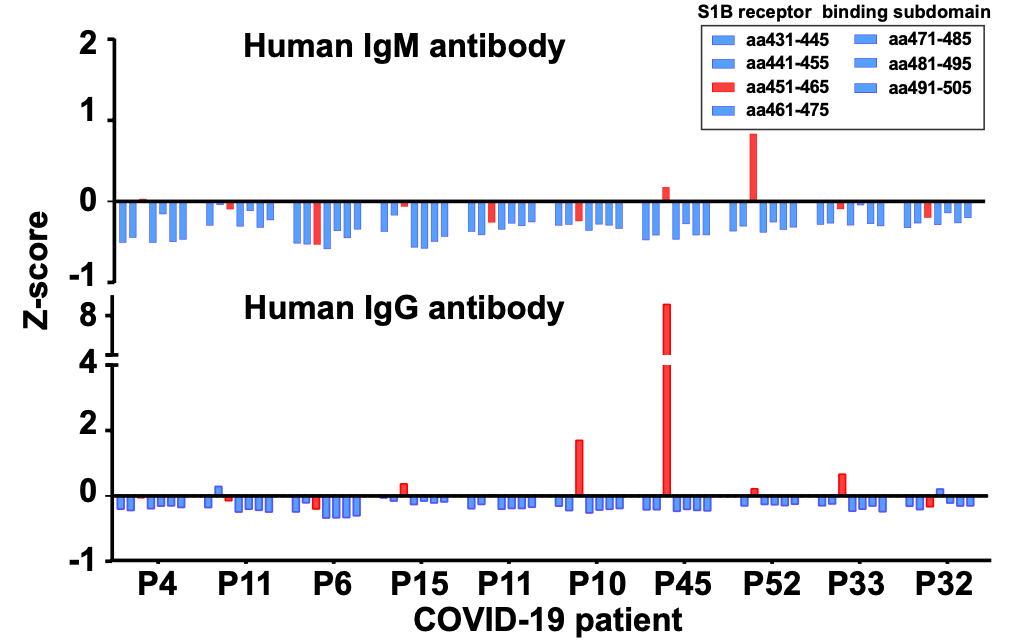
Figure 3: Antibodies against a potentially neutralizing epitope, shown in red, are observed to be enriched in multiple COVID-19 positive patients. Neutralizing epitopes are more commonly seen in IgG type antibodies, which arise later in the adaptive immune response.