Title: Spiky Nanostructures with Geometry-matching Topography for Virus Inhibition
Authors: Chuanxiong Nie, Marlena Stadtmüller, Hua Yang, Yi Xia, Thorsten Wolff, Chong Cheng, Rainer Haag
Journal: Nano Letters
In the past few months, mentions of ‘virus’ spiked across the internet and we all have learnt some new information about ‘virus’ in general. We know that this tiny organism can halt the world to a standstill. We know that it can affect the lives of more than 15 million people in just a couple of months. We also know that the researchers are working tirelessly now to develop a vaccine to fight against the virus that is responsible for the ongoing global pandemic.
But, are there other ways to combat the virus apart from the development of a vaccine? A team of researchers from Germany and China developed virus inhibiting agents by mimicking the geometry of virus with nanostructures. These nanostructures are in the comparable size regime and have identical shapes like the virus. Through geometry matching, nanostructures can attach to the virus and destroy the ability of the virus to attach to the hosts, i.e. human cells. In this way, virus infection can be prevented at the beginning.
In this work, researchers targeted influenza A Virus (IAV) which is responsible for influenza flu among birds and some mammals. To mimic the shape and structure of the virus, at first structure of the IAV virus was visualized (Figure 1a). Geometrically, it was assumed to be a sphere with a diameter around 100 nm with spikes on its surface with lengths of 10 nm. Nanostructures with similar shape were synthesized using nondegradable silica nanoparticles. All the nanostructures had a diameter around 150 nm, but the spike lengths were varied between 5, 10 and 30 nm (Figure 1b). A cartoon image of the interaction between the virus and the nanostructure is in Figure 1c.

The next question tackled here is – how does the spike length matter for the interaction between the virus and the nanostructures? A smooth nanostructure with no spikes did not show any binding to the virus which reinstated the idea of shape matching. Nanostructures with spike lengths of 5 and 10 nm showed favorable binding, but spike length of 30 nm showed relatively weaker binding. The researchers reasoned that longer spikes would present a gap between the nanostructure and the virus; hence, the surface area of contact will be minimized leading to a weaker binding which is explained in Figure 2.

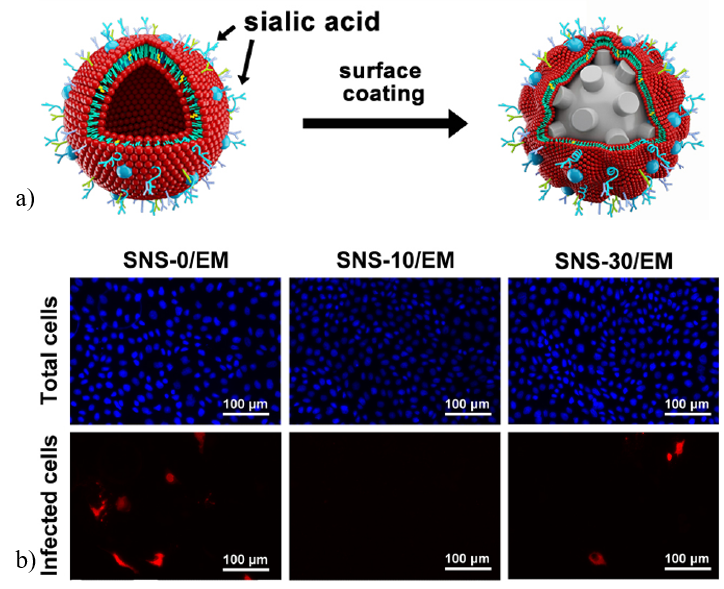
Although the virus particle was interacting with the nanostructures, but the virus was still infectious even when bound to the nanostructures. Hence, the nanoparticles were modified with a coating that mimics the host cells so that the virus can be tricked into attaching to the nanostructure. Generally, influenza A virus attaches to the host cell through sialic acid present in the host cells. Herein, sialic acid was coated onto these nanostructures and further studied for infection control (Figure 3a). Inhibition of cellular infection was visualized under a microscope. 10 nm spikes with sialic acid coating showed up to 85% inhibition as observed by no visible red colour in Figure 3b.
This study provides a new way of tackling viral infections apart from the well-established path of vaccines. As most of the viruses have spiky proteins attached to their core, these spiky nanostructures can be extrapolated to tackle other viruses too. Additionally, other geometric structures could also be explored to design new viral inhibitors. However, in vivo tests are needed to be done before actually using it on human and other mammals. But if it shows promising results, then this strategy has the potential to change the way viral infections are tackled in future.
Cover Image by PublicDomainPictures from Pixabay