Title: Multiplex Digital MicroRNA Detection Using Cross-Inhibitory DNA Circuits
Authors: Yannick Rondelez and Guillaume Gines
Journal: ACS Sensors
Year: 2020
https://pubs.acs.org/doi/10.1021/acssensors.0c00593
Featured image and figure reproduced with permission, and further permissions should be directed to the ACS.
We are all familiar with how electric circuits can be used to complete a job, such as a battery powering a light bulb to produce light. In this circuit, the battery supplies the input signal (stored energy) which is converted into a new signal (electric current) so that a light bulb produces the output signal (light). There is also likely a switch to turn the circuit on and off, as well as some kind of resistor to prevent too much current from entering the light bulb which could blow out the bulb. Electric circuits are so useful because they are made of modular components (batteries, wires, etc.) which can be recombined in different configurations to complete different tasks. In the past few decades, advances in biochemical technology have allowed scientists to start creating biochemical circuits, where biological molecules—such as the two kinds of nucleotide strands: DNA and RNA—act as the modules to complete a task, such as amplifying a molecular signal in a biological sample to high levels for accurate and quantitative detection.
Recently, Rondelez and coworkers have been designing a biochemical circuit for the detection of microRNAs which are very short nucleotide strands made by the cell to regulate how genes are expressed into functional cell components like enzymes. Dysregulation of microRNAs (when the cell makes too many or too few) has been linked to diseases like cancer, diabetes, neurological disorders, and heart disease. Therefore, detecting the amount of a certain microRNA in a patient could be used to diagnose disease or track the progress of a patient’s treatment. Conveniently, these microRNAs can be found in blood or urine. However, the microRNAs exist at very low concentrations in these samples, making it difficult to accurately detect the amount present [1]. Therefore, Rondelez and coworkers decided to design a microRNA-sensing circuit that would amplify the signal from a low concentration of microRNA.
The basis for their circuit is the Polymerase Chain Reaction (PCR) where an enzyme called DNA polymerase reads a template DNA strand and builds a new DNA strand with the complementary nucleotide sequence, such that the nucleotide adenine (A) pairs with the nucleotide thymine (T), and guanine (G) pairs with cytosine (C). For example, if the template strand is ATGCTTC, the complementary strand would be TACGAAG. Because DNA polymerase can complete this process many times, one template strand can be used to make many complementary strands, which will be useful in the biochemical circuit to create a large output signal from a small number of microRNAs.
Importantly, however, DNA polymerase cannot start a new DNA strand from scratch; instead it must extend an existing nucleotide strand. In PCR, a very short nucleotide strand called a primer is complementary to one end of the template strand, and this primer is used to initiate PCR. The microRNA of interest (i.e. that the scientists want to detect) acts as the primer for the first template. Therefore, the microRNA is the switch of the circuit: when it’s present, the circuit should be turned on. When it’s absent, the circuit should be turned off.
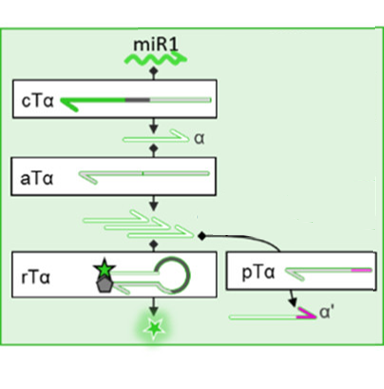
The scientists designed their microRNA-sensing circuit (Fig. 1) using four different templates to perform different functions in the circuit. First, the “conversion template” converts the signal from the microRNA sequence itself (bright green) into a generic DNA sequence called an “amplicon” (light green) that will be used in the rest of the circuit. An enzyme called “nickase” cuts the DNA at a specific site in the sequence—similar to how scissors cut a piece of paper—to separate the amplicon from the original microRNA. Overall, this conversion step ensures that if the researchers want to repurpose their circuit to detect a different microRNA with a different nucleotide sequence, only the conversion template will have to be redesigned, leaving the other three templates unchanged.
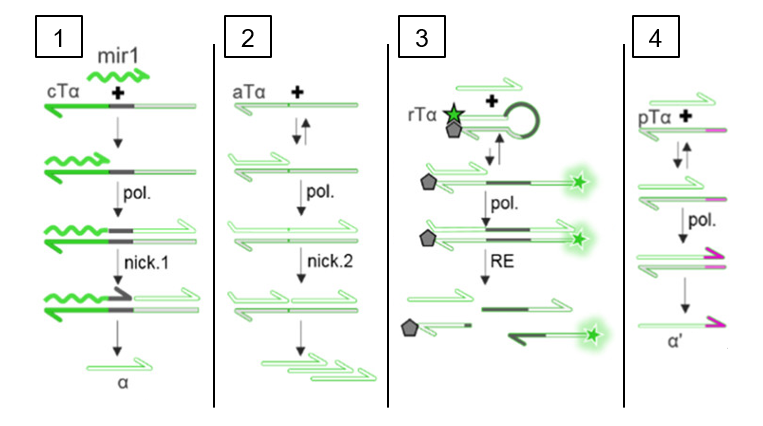
Second, the “autocatalytic template” amplifies the signal by making more copies of the amplicon. Thirdly, the signal is converted into fluorescent light using the reporter template. The reporter template has a fluorescent molecule bound to one end and a quencher molecule that prevents fluorescence bound to the other. Initially, no fluorescence is produced because the quencher and fluorescent molecule are too close together. However, PCR of the reporter template creates a new sequence (dark green) that can be recognized by an enzyme called a “Restriction Enzyme” which cuts the DNA at a specific site (similar to the nickase in step 1). This separates the quencher from the fluorescent molecule so that green fluorescent light can be detected. This is a positive test result that the microRNA of interest was indeed present in the sample.
However, in a real reaction mixture, PCR can be “leaky,” meaning that if enough time passes, PCR and eventually fluorescence will be seen even in the absence of the original microRNA. The almost inevitable nature of false positives would make this biochemical circuit useless for tracking disease states, but a fourth module can be added to the circuit to solve this problem. In this module, a pseudotemplate (Fig. 1, light green and pink) is used to add a few nucleotides (pink) to the end of amplicon (light green). This modified amplicon can no longer be used by the autocatalytic template or the reporter template, so the leak does not result in a false positive. Only a small number of pseudotemplate strands are added, so that a real signal initiated by the microRNA of interest will make enough amplicon to far surpass the pseudotemplates present. This leads to a strong fluorescent signal that’s proportional to the number of microRNA strands in the original sample.
With their biochemical circuit, Rondelez and coworkers can successfully quantify the amount of a specific microRNA in a sample. More experiments are still needed, however, before this system can be used at a doctor’s office to track microRNA dysregulation in a patient. For example, a blood or urine contains many different microRNAs and other biological molecules which could possibly disrupt the circuit, and additional purification steps may be required. For now, however, this circuit serves as an important step toward the future goal of using microRNA dysregulation as a marker to diagnose and track disease.
[1] Gines, G.; Menezes, R.; Nara, K.; Kirstetter, A.-S.; Taly, V.; Rondelez, Y. Isothermal Digital Detection of MicroRNA Using Background-Free Molecular Circuit. Sci. Adv. 2020, 6, eaay5952. DOI: 10.1126/sciadv.aay5952