Title: Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies
Authors: Jiahui Chen, Kaifu Gao, Rui Wang, and Guo-Wei Wei
Year: 2021
Journal: Chemical Science
There can be no denying, the COVID-19 pandemic is one of the biggest global health crises the current generation has had to face. Currently, the most promising strategy to combat the raging pandemic is through a global vaccination strategy with many countries already boasting early victories. However, it hasn’t all been a linear path to success. Emerging variants of the virus are producing more contagious strains and speculation is growing about what these mutants may mean for the efficacy and reliability of our current vaccine strategies. Scientists are already beginning to address these concerns with a study published last month which aimed to predict how mutations of the SARS-COV-2 virus may threaten COVID-19 vaccines and antibody therapies.
So how do vaccines work to help protect us from pathogens? Many vaccines aim to elicit an immune response from the human body, with many (including COVID-19 vaccines) doing this by encouraging the body to generate antibodies in response to the vaccination without causing disease. This means that when exposed to the pathogen, your body already has the tools required to fight off infection. So, what actually are antibodies and how do they work? Antibodies are highly specialised proteins that bind structures called antigens found on the surface of foreign invaders i.e. viruses. Through this binding, your immune system can then identify these invaders as foreign and thus neutralise them. Antibodies exist as a part of the adaptive immune system and are highly specialised to each specific antigen that you may encounter. Since this part of the immune system is ‘adaptive’, it works most effectively when it relies on immunological memory to fight off foreign invaders that it has already been exposed to before. Therefore, to defeat COVID-19, it is imperative that we have current and reliable vaccine strategies.
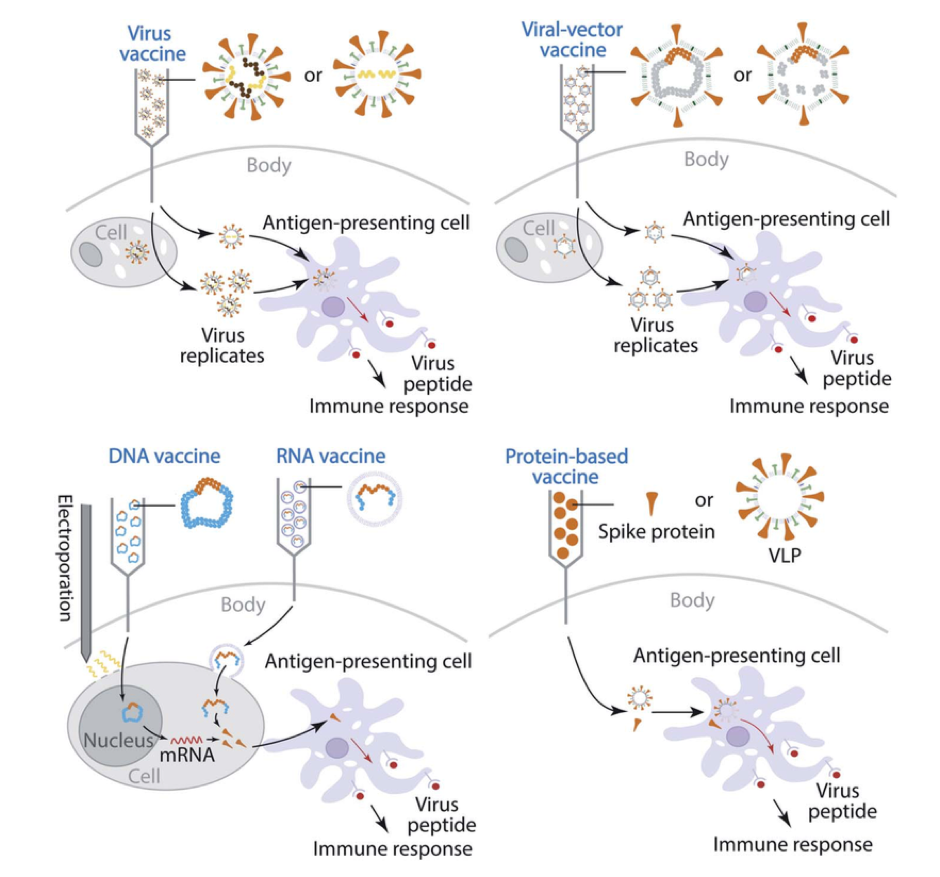
There are four vaccine strategies for COVID-19 currently available or in development (Figure 1). In all cases, non-infectious viral fragments are taken up by antigen-presenting cells where large proteins are broken down into small viral peptides and presented on the surface of the cell to elicit an immune response. The way these vaccine strategies differ, is how and what viral particles are presented to the human body through injection. They may be in the form of weakened or inactivated viruses, viral vectors (AstraZeneca Oxford vaccine), or viral DNA or RNA (Pfizer and Moderna vaccines) which all use our cells machinery to produce viral surface proteins for antibody recognition. Additionally, there is a protein based vaccine approach which aims to inject viral proteins directly into the body. Many COVID-19 vaccines currently available and in clinical trials target antibody binding to the spike protein (found on the surface of the virus) to neutralise the pathogen. However, there is growing concern that if mutations occur, particularly located on the spike protein, this may reduce the effectiveness of some vaccines.
So what does it mean when a virus becomes mutated? Mutation occurs during the process of replication, where a virus hijacks your cell machinery to make more copies of itself. Mutation occurs because of mistakes in replication; where specific points of the viral genomic sequence differ from its immediate predecessor. This may sound like a bad thing for the virus, however, occasionally mutations occur that result in a more successful pathogen, giving us new variants of COVID-19 i.e. the B.1.1.7 strain contains mutations causing it to become highly contagious. This becomes concerning when mutations occur on regions of the virus that are important for antibody recognition, such as on the spike protein, which may result in antibodies not being able to bind and neutralise the virus anymore thus rendering current vaccines ineffective. Therefore, research into likely mutations and how they may affect vaccines, such as that conducted in the recent study by Chen and co-workers, becomes imperative.
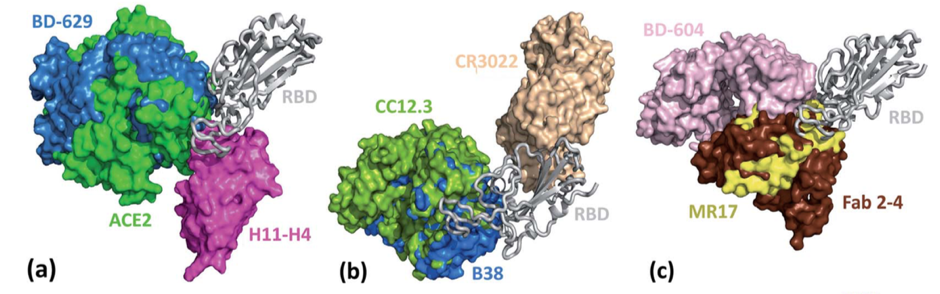
To conduct their study, the authors retrieved more than 200 000 complete SARS-COV-2 genome sequences from the GISAID data base (an open access genome data base of influenza and SARS-COV-2 viruses). Using these genomes, they identified more than 26 000 unique single mutations and additionally computed the frequency at which the mutations appeared in each sequence. From this, they identified that there were 5003 unique single mutations that existed on the spike protein alone. Next, the authors constructed a library of 56 SARS-COV-2 antibodies (Figure 2) by accessing the existing structures from the Protein Data Bank (PDB; a data base for the 3D structure of large biomolecules).
To study the potential impacts of mutations of the SARS-COV-2 spike protein, the authors set out to evaluate the binding free energy (BFE) differences between mutated spike proteins and their antibody complexes compared with the parent SARS-COV-2 strain. The BFE (ΔG) of a protein-protein complex is a measure of how energetically favourable a bound protein is when compared to the unbound protein with a more negative ΔG value indicating more favourable binding. The authors used computational modelling to determine the change in BFE from before and after virus mutation to gauge how the mutations may affect antibody binding. The change in BFE (ΔΔG) is denoted as ΔΔG = ΔGW – ΔGM; where ΔGW is the BFE of the wild type SARS-COV-2 spike protein and ΔGM is the BFE of the mutant. Therefore, a negative ΔΔG value means that the mutant has resulted in a less stable protein–protein interaction which results from a larger ΔGM value than the parent wild-type SARS-COV-2 binding.
Overall, the authors found that most mutations of the SARS-COV-2 spike protein resulted in a weakened binding interaction with antibodies that bind to the spike protein, with a total rate of 71% of mutations causing negative BFE (ΔΔG) changes. With such a high percentage of mutations causing weakened binding interactions, this research has highlighted the vulnerability of our current vaccine strategies. It is clear from these findings that the development of mutation-resistant vaccines will be imperative for the future and that we may need to prepare for seasonal vaccinations similarly to what is carried out for the influenza virus.
In addition to spike protein–antibody complexes, the authors also examined the change in BFE for spike protein complexes with angiotensin-converting enzyme 2 (ACE2). The ACE2 protein is found on the surface of many of our human cells and serves as an entry point into our cells for the SARS-COV-2 virus through initial binding with the spike protein (Figure 2). Interestingly, the authors found that whilst most mutations caused a weakened binding of the spike protein–antibody complexes, many mutations (65%) produced a positive BFE change for the spike protein receptor binding domain with ACE2. This indicates that many mutations, including those seen in the UK and South African variants of the virus will strengthen the binding between the virus and the entry point into human cells resulting in more infectious strains of the virus.
Through this research, the authors have provided methods to predict how specific mutations may influence the efficacy and reliability of certain vaccines; a tool which may prove invaluable leading into the future of the pandemic. The use of open-access data bases such as the GISAID and the PDB has allowed these researchers to highlight the current vulnerability of many vaccines to emerging and future variants accentuating the pressing need for more mutant resistant treatments and the potential for seasonal vaccinations. The current research into the effects of mutations on COVID-19 vaccines will only be improved and made more reliable when more antibody structures become available.