Featured image (above) and all figures (below) are reprinted with permission from Gotrik et. al, JACS, 2018, 140, (10), 3583-3591. Copyright 2018 American Chemical Society.
Title: Direct Selection of Fluorescent-Enhancing RNA Aptamers
Authors: Michael Gotrik, Gurpreet Sekhon, Saumya Saurabh, Margaret Nakamoto, Michael Eisenstein, and H. Tom Soh.
Journal: Journal of the American Chemical Society: https://pubs.acs.org/doi/10.1021/jacs.7b10724
Year: 2018
Imaging biomacromolecules is of paramount importance in chemical biology. Genetically encoding an imaging “tag” to a biomacromolecule of interest is currently one of the leading ways to observe cellular components in real time. This strategy works particularly well for proteins as this tag can often be seamlessly fused to the protein’s amino acid sequence by simply adding the right codons to its DNA sequence. Green fluorescence protein (GFP), for instance, can be attached to a multitude of targets and has become an indispensable tool to elucidate in vivo processes where these proteins participate in. Despite the proficiency in protein labeling, the genetic encoding of imaging tags for RNA for imaging is still particularly challenging.
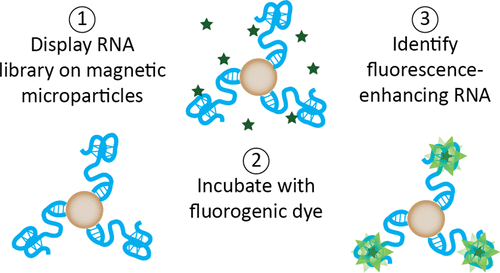
In this account, the lab of Tom Soh at the Chemical Engineering department at Stanford seeks to address this limitation through the development of a method to find and characterize RNA molecules, known as aptamers, that bind to a particular ligand and fluoresce upon such binding. The backbone of this research is the ability to identify suitable aptamers that (a) bind well to the small-molecule ligand, (b) enhance the ligand’s fluorescence properties, and (c) operate efficiently both in vitro and in vivo. We can now discuss how exactly the group conceived a method that complies with all three requirements.
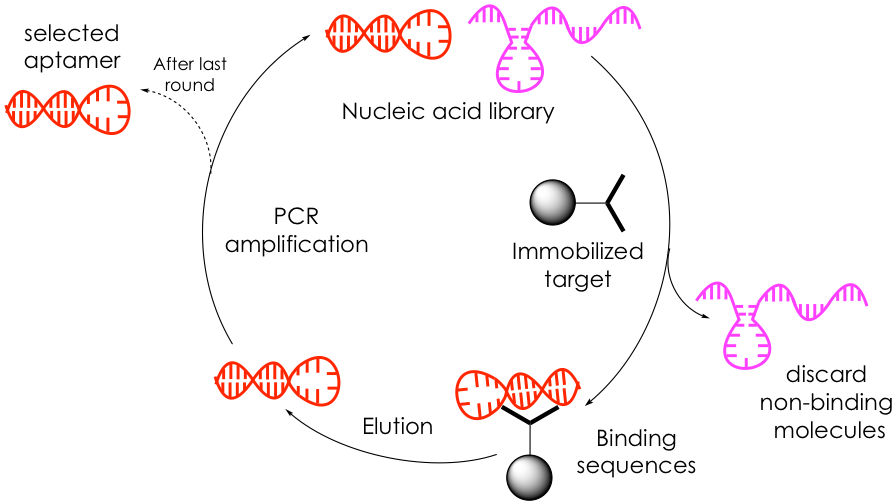
Aptamers are typically discovered through a process known as SELEX, which consists of repeating rounds of binding, partitioning, and amplification of a large library of randomized, short nucleic acids (with up to 1016 members!) to find sequences that most tightly bind to a target (Figure 1). The process begins with the incubation of the target molecule, often immobilized to a bead, with the nucleic acid pool. Unbound oligonucleotides are discarded via rigorous washing steps while bound sequences are selectively dissociated from the substrate and collected. This reduced, yet promising library is amplified through polymerase chain reaction (PCR) to generate an enriched oligonucleotide pool that is used for the next round of selections. The binding-partitioning-amplification sequence is repeated ~20 times before a small group of highly affine target-selective aptamers is isolated and characterized by DNA sequencing to determine their base sequence.
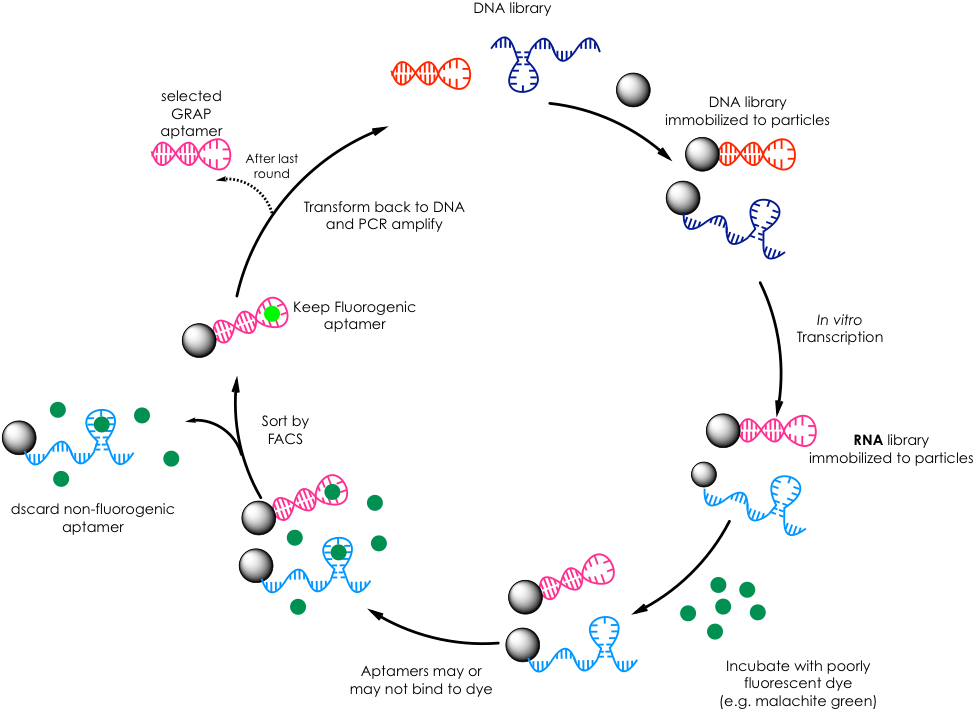
The Soh lab borrows from this technique but adapts it not only to find RNA aptamers that bind to a particular dye molecule but that, upon such binding, enhances the fluorescence of said dye, thus elevating SELEX to a bimodal approach (Figure 2). First, they immobilize a randomized DNA library to very small beads or particles such that each particle contains a single copy of each unique DNA sequence. Then they transcribe the DNA to RNA, and they expose these RNA-containing particles to a dye of interest that doesn’t fluoresce in solution such as malachite green. Subsequently, they use a technique known as FACS that rapidly analyzes each particle and sorts it based on fluorescence. Fundamentally, FACS excites the particle-RNA-dye complexes and looks for a fluorescence response that exceeds a threshold. Particles whose RNA-dye interactions were able to generate enough signal are physically separated from those that failed to meet the threshold. These promising particles are then amplified by PCR and subjected to more rounds of selection. After performing several rounds, the group was able to reduce the library to a handful of high-fitness aptamers, which were finally identified by sequencing. The group termed this methodology GRAP (gene-linked RNA aptamer particles) display.
To extend their proof-of-concept, the authors then chemically synthesized each RNA aptamer and performed more thorough fluorescence and affinity titrations with malachite green. They also compared the properties of their aptamers to those of previously developed malachite green aptamer. Delightfully, some GRAP aptamers not only showed greater affinity and fluorescence enhancement of malachite green than the pre-existent aptamer, but they also displayed diverse spectral properties whit very different excitation and emission wavelengths. The curious reader is encouraged to refer to the original paper for more detailed optimization and assay metrics.
The question now arises of how exactly can this technology be incorporated into living systems to image RNAs of interest. One could envision fusing the sequence of one of the GRAP aptamers to that of a biologically relevant RNA and then supplying the cell with malachite green such that the tagged species fluoresces. The authors are still far from achieving this goal in this particular paper, but the solid proof-of-principle they cemented is laudable nonetheless. In this regard, only time will tell if GRAP can be readily extended as a robust tool for fluorescent RNA sensors.