Title: Post-Translational Tyrosine Geranylation in Cyanobactin Biosynthesis
Authors: Maho Morita, Yue Hao, Jouni, K. Jokela, Debosmita Sardar, Zhenjian Lin, Kaarina Sivonen, Satish Nair, and Eric W. Schmidt.
Journal: Journal of the American Chemical Society
Year: 2018
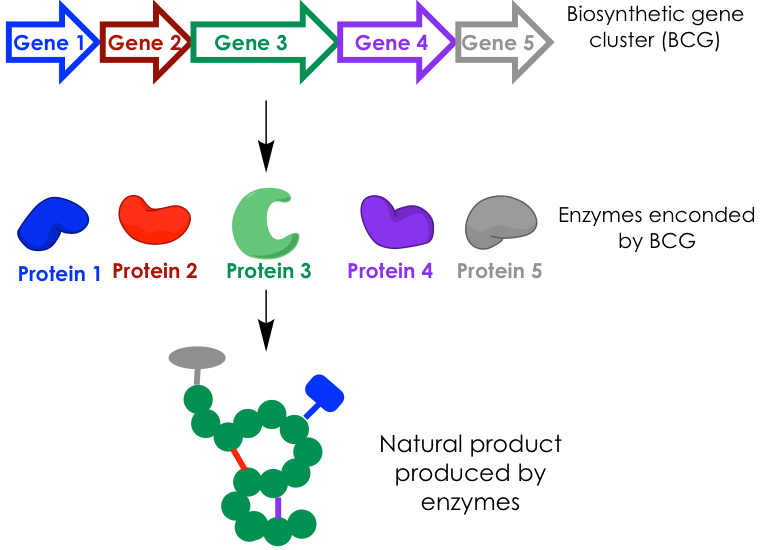
Microorganisms are true synthetic powerhouses. It is remarkable how relatively simple microbes and fungi can generate an overwhelming collection of small molecules, known as natural products, which possess incredible architectural and functional diversity. The components responsible for this ability are portions of the microorganismal genome known as biosynthetic gene clusters (BCGs), usually represented by a line of arrows. These are stretches of contiguous genes that code for a collection of proteins, each in charge of a specific step in the biosynthesis of a natural product. If we can think of this synthesis like an assembly line in a factory, we can compare the proteins to the machines that piece together the target good and the BCGs to the blueprints of that assembly line (Figure 1).
Out of the many classes of natural products, some bacteria are very good at generating ribosomally-synthesized and post-translationally modified peptides, or RiPPs for short. While the ribosome is mostly known for translating full-size proteins out of messenger RNA, it can also make smaller peptides of relatively simple sequence that are later modified by tailoring enzymes encoded by the BGCs to make the mature RiPP product. These chemical styling steps are necessary because they impart the RiPP core with new moieties that grant the RiPP with antimicrobial, antifungal, surfactant, or signaling function.
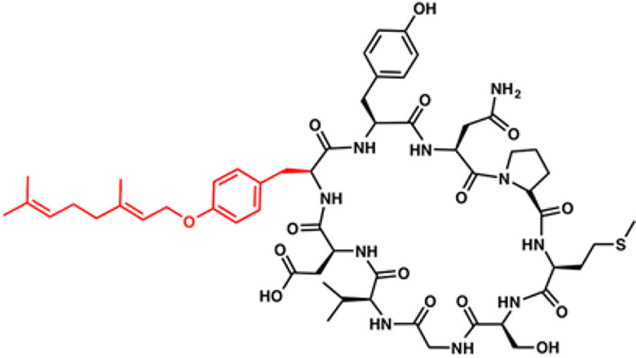
In this paper, researchers from the University of Utah and the University of Illinois focus their attention on a class of RiPP known as a cyanobactin. Cyanobactins are small peptides whose N- and C-termini have been stitched together to form a macrocyclic peptide. On top of that, they often also display several modifications, such as methylations, heterocyclization of cysteine or serine residues, or additions of 5-carbon units (known as prenylation) to serines, threoni
nes, or tyrosines.
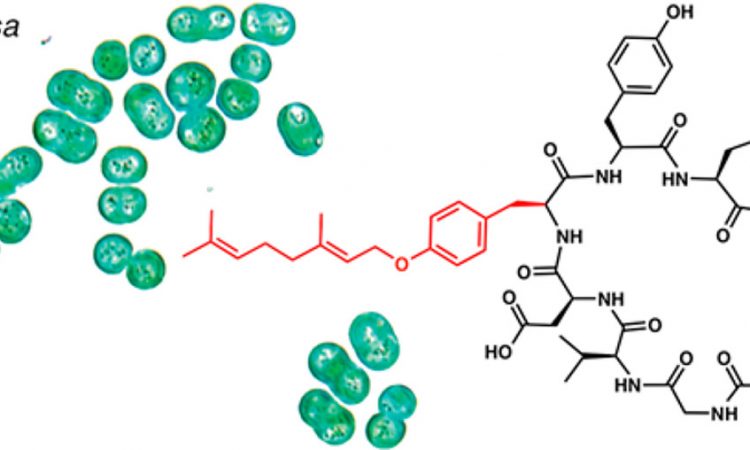
The authors became particularly interested in a cyanobactin known as piricyclamide (Figure 2) when they observed that the mass spectroscopy signature of this molecule suggested that a tyrosine was modified with a 10-carbon fragment. This event, also known as tyrosine geranylation, is an unprecedented post-translational modification of RiPPs.
The authors started by doing some bioinformatics analysis on the genome of the freshwater cyanobacterium M. aeruginosa in search of the BGC responsible for making piricyclamide. Once they found, they hypothesized that the enzyme PirF might catalyze the transformation of interest. They purified the protein and first reacted it against isolated amino acids (serine, threonine, and tyrosine) along with a geranyl source. After analyzing the reaction by chromatography coupled to mass spectrometry, they found that PirF selectively modifies tyrosine with the geranyl group. They then moved to analyze the substrate scope of PirF and found that it can functionalize cyanobactins of diverse structures, suggesting that other cyanobactins might also contain this modification.
The authors continue to show validations that PirF indeed catalyzes this transformation selectively on tyrosines with a broad overall RiPP promiscuity. While the role of tyrosine geranylation and the exact mechanistic details of its installation are not explored just yet, this work marks the discovery of an unappreciated RiPP modification that could be more abundant and possess interesting biochemical roles.