Article title: New codons for efficient production of unnatural proteins in a semisynthetic organism
Authors: Emil C. Fischer, Koji Hashimoto, Yorke Zhang, Aaron W. Feldman, Vivian T. Dien, Rebekah J. Karadeema, Ramkrishna Adhikary, Michael P. Ledbetter, Ramanarayanan Krishnamurthy & Floyd E. Romesberg Lina M. González, Nikita Mukhitov and Christopher A. Voigt
Nature Chemical Biology (2020)
What makes an organism synthetic vs. non-synthetic? It is generally agreed that natural, or non-synthetic, organisms use the same genetic code and protein-building hardware that evolved billions of years ago. This means that natural organisms are limited to the 4 chemical bases that make up DNA and RNA and the 22 amino acids that make up proteins. However, researchers have shown in the past few decades that is it possible to expand these capabilities. There are two primary ways to do this: engineer organisms to use more than the 4 natural DNA/RNA bases or engineer them to use additional amino acids – beyond the 22 that can naturally go into proteins. The researchers here show for the first time that both can be accomplished in a single organism!
For some background, proteins provide the structures and tools for biological function. Imagine proteins as the hammers, fences, bricks, and trucks for the biological world – providing structure but also function. All proteins are composed of amino acid building blocks, with different amino acid combinations leading to different classes and functions of protein. To code for all these proteins, the organisms maintain a source code, DNA.
The code of life, DNA, has 4 different chemical bases that code for everything in the cell: A, T, C, and G. Think of this code as a recipe. This code is then read by the chef of the cell, the ribosome, 3 at a time, e.g. ATC, TAT, GAT. These clusters of three bases are known as codons and each one codes for a certain amino acid, which are the ingredients of the recipe. How many different codons can these natural 4 bases make? Some simple math, 4^3, gives us a value of 64. You might be asking, why can’t cells code for 64 amino acids then? The genetic code is somewhat redundant, meaning that a single amino acid can have multiple codons that code for it. This is just how life naturally evolved and it may be helpful to think of these multiple codons as different brands of the same ingredient.
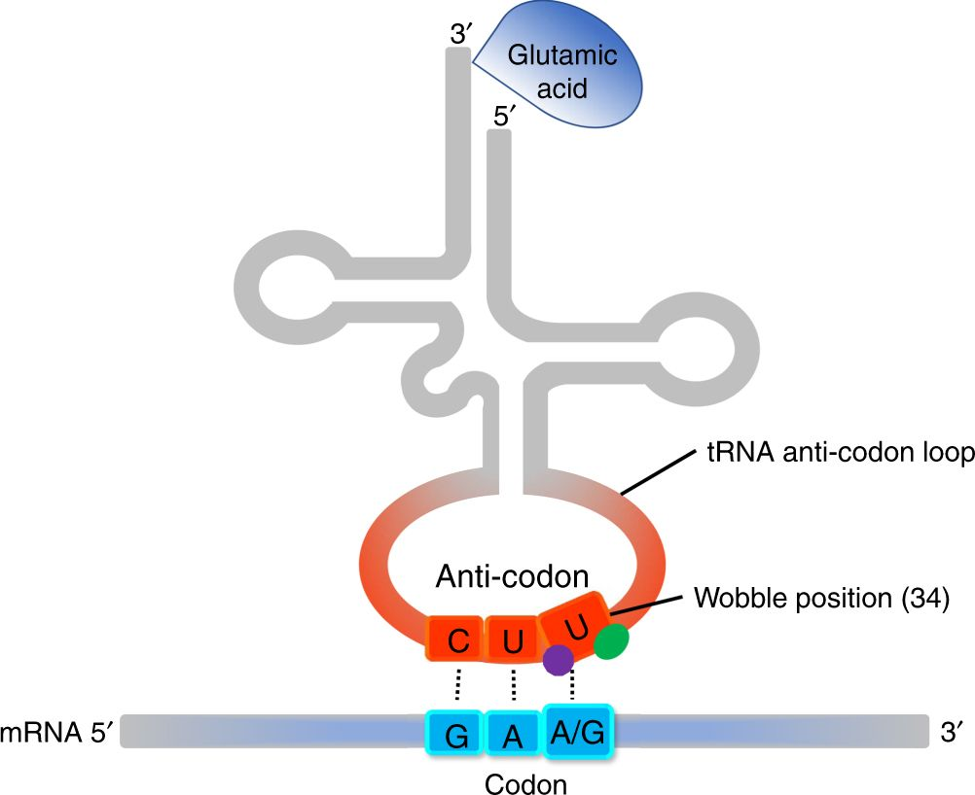
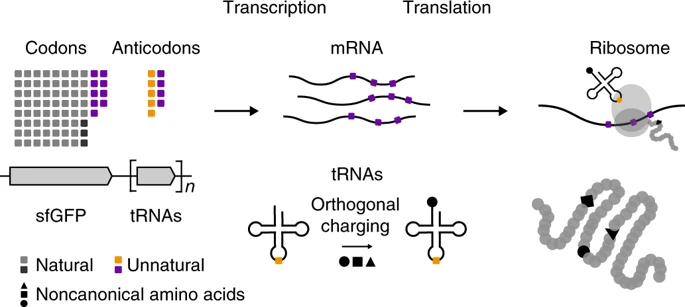
Another piece of cellular machinery, known as a transfer RNA (tRNA), delivers the amino acid to the ribosome to generate each piece of the protein (Figure 1). Notice the codon at the bottom on the mRNA strand, the anti-codon that recognizes it and the amino acid that is attached at the top. Everything must work precisely for the amino acid to be attached to the growing protein chain. Once the ribosome reaches a codon that tells it to stop, the recipe is finished and the protein floats away to perform its function. A full overview is shown in Figure 2, where the synthetic codons are shown in purple. These synthetic codons can then code for unnatural amino acids (shown as black shapes) to make never-before-seen unnatural proteins.
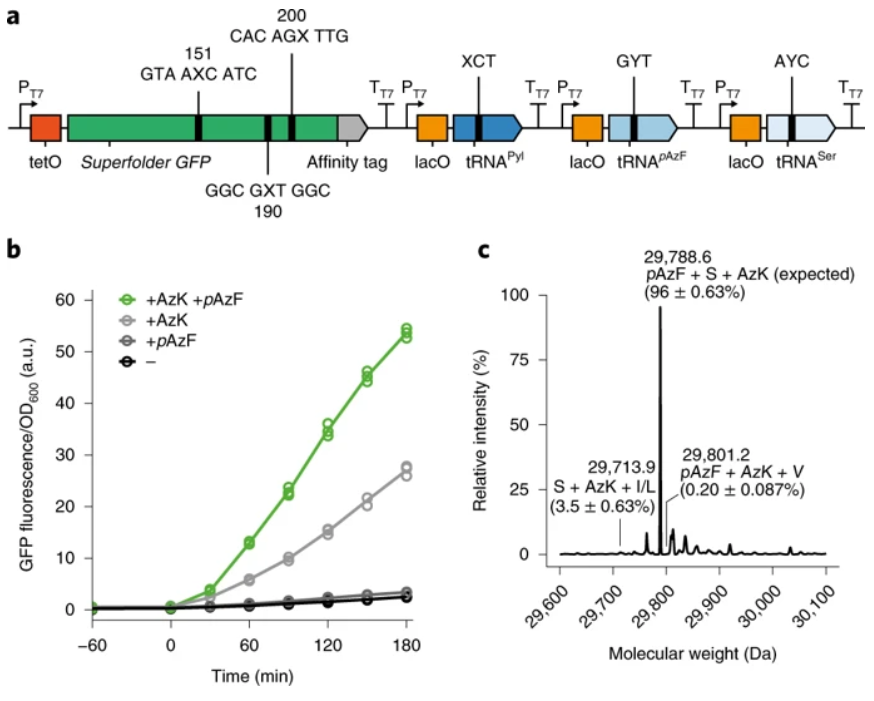
To expand beyond the 4 chemical bases of DNA, the researchers used 2 unnatural bases that they had developed in a previous study. Instead of A, T, G, or C, these were labeled X and Y. X and Y pair with each other, like how A pairs with T and C pairs with G in natural DNA. They then created randomized sequences of codons to code for specific amino acids in a fluorescent protein called GFP (green fluorescent protein), meaning that the codon could be ATX or CGY or any other combination of three. Usually, mutating a protein by changing an amino acid like this will affect the function so the researchers chose specific spots that are known to not break the protein. To find the synthetic codons that worked, the researchers looked for cells in which the green fluorescent protein was made properly and glowed green which gives a simple visual confirmation when the protein is translated. Using this method, the researchers found a handful of combinations that worked.
The researchers go on to show that 3 out of the 9 of these new codons they’ve generated are what’s called orthogonal – meaning they do not interfere with the code of each other or the native code that’s already in the organism. The other 6 you can think of like small bugs in a programmer’s code, they mess up the rest of the code because they are not performing their function correctly – instead they are incorporating amino acids that they weren’t designed to code for. The three orthogonal codons can be used together without causing any significant bugs in the code. The researchers then took the 3 new orthogonal codons and showed that they could together be used to generate GFP with unnatural amino acids called AzK and pAzF (Figure 3).
This is the first time that “unnatural” codons have programmed “unnatural” proteins, meaning that the researchers completed the entire coding-to-protein generation cycle using both synthetic code and synthetic hardware. This work moves beyond what is capable in natural organisms, into a world of semi-synthetic organisms and perhaps eventually, fully synthetic organisms.
The research here showcases a semi-synthetic organism that can code for proteins using 67 instead of the standard 64 codons. Although no groundbreaking applications are shown, the researchers lay the foundation for programming living organisms which may carry out entirely new functions. One can envision a multitude of new functions from sensing to chemical synthesis so keep an eye out for more innovation in the future.

