Title: Spreading of a virulence lipid into host membranes promotes mycobacterial pathogenesis
Authors: CJ Cambier, Steven Banik, Joseph A. Buonomo, Carolyn Bertozzi
bioRxiv: June 2020
doi: https://doi.org/10.1101/845081
The current pandemic has shown how deadly viruses can be, but bacterial infectious diseases can be equally devastating. Notably, tuberculosis, a pulmonary disease caused by the bacterium Mycobacterium Tuberculosis, kills more than a million people every year, predominantly in developing countries. It is airborne and highly contagious since a small number of bacteria in tiny droplets in the air can infect a person and incubate in their body for months before the onset of symptoms1.
This disease has affected humans for thousands of years, during which mycobacteria have evolved to be able to circumvent the immune system. Normally, pathogens activate an immune response by being detected by toll-like receptor (TLR) proteins, which are found in the surface of immune cells. Think of TLRs as ID detectors, they bind to molecules and can detect whether these are native to the body or foreign to it. If they find that they are foreign, then they activate a response to alert the immune system of the presence of a pathogen. Specialized white blood cells are then recruited to that site, which swallow and destroy the pathogen through a process called phagocytosis. However, mycobacteria do not activate this immune response, which allows them to transfer into red blood cells where they can incubate for long periods of time while they replicate.
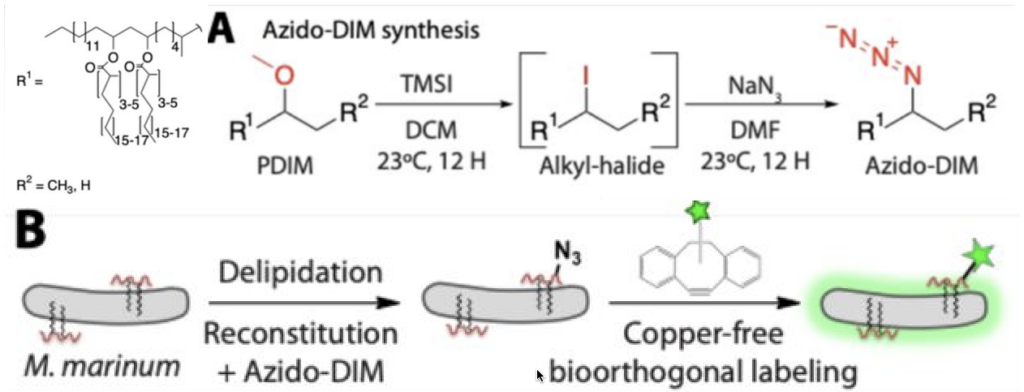
The process by which mycobacteria are able to avoid detection by TLRs is not well understood, but an interesting clue arose in a study performed previously by the lead author of this paper. They used genetic engineering to develop a strain of mycobacteria that is unable to synthesize phthiocerol dimycocerosate (PDIM), which is a lipid found in the outer envelope of the cell wall. When zebrafish were infected with this strain of mycobacteria, the TLR response was activated, and white blood cells were able to kill the mycobacteria. This suggested that PDIM may play a role in inactivating the TLR response, but the results were not conclusive because genetic alterations can affect the composition of many lipids in the outer cell wall, and not just the targeted one.
To track PDIM throughout the course of infection of zebrafish with mycobacteria, the Bertozzi group developed a method to tag the lipid with a fluorescent probe. They first removed the native PDIM by using extraction with non-polar solvents. Since lipids are highly non-polar, immersing the cells in these solvents would remove the PDIM from the outer cell wall because the lipids would be more attracted to the solvent. Once PDIM was extracted, they switched an ether group(ROR’) to an azido group (N3) in a two-step synthesis. The modified PDIM were then added back to the mycobacterium. Azido groups react readily with strained alkenes in water, allowing PDIM to be functionalized with a fluorescent label while bound to the outer envelope (Figure 1). Reactions like this one are called “bio-orthogonal” because they can be carried out in vivo without interrupting biological processes.
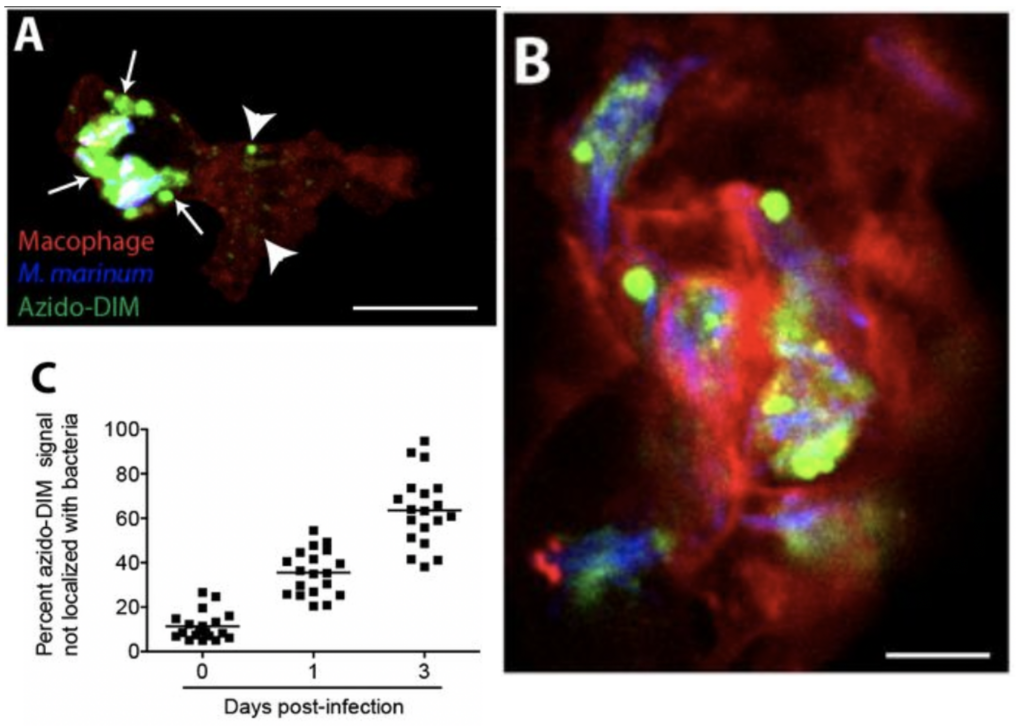
Mycobacteria with the fluorescently tagged PDIM were then used to infect zebrafish. Since this model organism is transparent, they were able to detect the location of PDIM with microscopy. They found that PDIM spreads away from the mycobacteria and into the zebrafish’s white blood cells as the infection progressed (Figure 2). As a control, they also performed the same experiment with a fluorescently tagged generic lipid and found that it remained bound to the mycobacteria during the course of the experiment, showing that the mechanism of spreading is unique to PDIM. They also developed a method to increase the rigidity of the outer envelope of mycobacteria by immersing them in paraformaldehyde. They found that this decreased the spread of the fluorescently tagged PDIM away from the outer envelope during infection. In this case, the TLR-dependent immune response was activated, and the bacteria were killed by white blood cells. These experiments reveal that PDIM is able to spread into human cells and is essential to the inactivation of the immune response. However, more studies are needed to understand how it is able to accomplish this.
This study is a great example of how chemistry can be leveraged to make tools that are useful for studying biology; chemical methods allowed for the introduction of a fluorescent tag to PDIM, which uncovered its role during infections by mycobacteria. The paper’s findings also illustrate the complicated mechanisms pathogens evolve to be able to survive inside their hosts, and why tuberculosis is still a major issue affecting humanity in the present.
- “Global Tuberculosis Report” (PDF). WHO. WHO. 2019. Retrieved 24 March 2020.