Title: Photochemical Identification of Auxiliary Severe Acute Respiratory Syndrome Coronavirus 2 Host Entry Factors Using μMap
Authors: Suzuki, S., et al.
Journal: Journal of the Academy of Chemical Sciences
Year: 2022
DOI: 10.1021
Photo by Fusion Medical Animation on Unsplash
SARS-CoV-2 attaches to human cells and initiates infection by binding its spike protein to ACE2 receptors that are highly expressed in the surface of cells of the respiratory system (Figure 1). This interaction was discovered at the beginning of the COVID-19 pandemic, but there are still details about how the virus initiates infection that remain unclear. Notably, ACE2 is expressed at very low levels throughout most human cells, while COVID-19 can affect multiple organ systems. Could other receptors interact with the spike protein and facilitate the infection of cells outside the respiratory system?
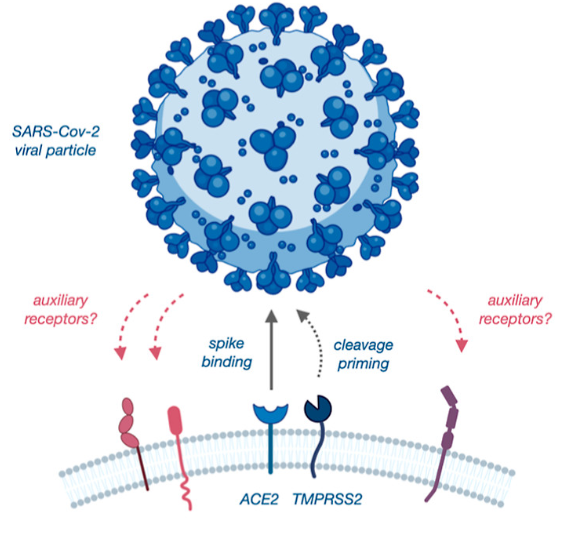
Figure 1 – Mechanism of action for the initiation of infection by SARS-CoV-2. Reprinted with permission from J. Am. Chem. Soc. 2022, 144, 36, 16604–16611. Copyright 2022 American Chemical Society
To answer this question, Suzuki et al. used proximity labeling, a strategy in which molecules close to a protein of interest are tagged with a chemical label. This label facilitates the purification and identification of molecules that interact with the protein of interest, which can be important for deconvoluting the mechanism of action of a biological process. They used a novel proximity labeling technique called µMap, in which the protein of interest is conjugated with an iridium catalyst molecule and is incubated with a biotin-diazirene compound (Figure 2). When irradiated with blue light, this molecule can catalyze a reaction in which the biotin-diazirene releases an N2 molecule and leaves behind a biotin-carbene. The carbene’s lone pair quickly undergoes an insertion with nearby C-H bonds, thus forming a covalent bond between biotin and nearby organic molecules. These molecules can be isolated by using streptavidin, which forms a strong interaction with biotin, and then be identified through mass spectrometry-based techniques.
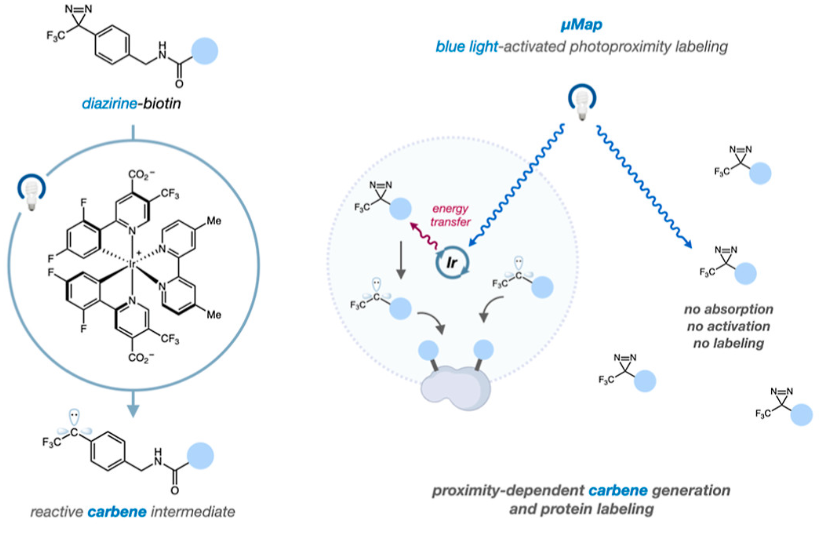
Figure 2 – (Left) Depiction of the diazarene-biotin warhead molecule before and after the reaction catalyzed by the iridium. (Right) The steps involved in the µMap technique. Reprinted with permission from J. Am. Chem. Soc. 2022, 144, 36, 16604–16611. Copyright 2022 American Chemical Society
To identify novel receptors that interact with SARS-CoV-2, they incubated iridium catalyst tagged spike protein with human lung cells and carried out the µMap technique (Figure 3). After isolation using streptavidin and mass spectrometry, they identified 8 receptors that had been tagged through proximity labeling.
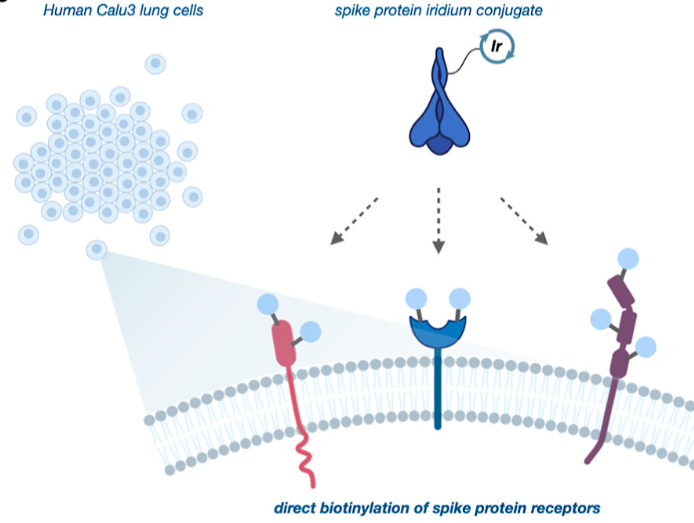
Figure 3 – SARS-CoV-2’s spike protein was conjugated with the Iridium catalyst and incubated with human lung cells and biotin-diazirene molecules. Irradiation with blue light enabled the biotinylation of receptors that could be interacting with the Spike protein. Reprinted with permission from J. Am. Chem. Soc. 2022, 144, 36, 16604–16611. Copyright 2022 American Chemical Society
To confirm that these 8 receptors can interact with the spike protein of SARS-CoV-2, the authors created pseudoviruses that express the spike protein on their surface. These pseudoviruses contain RNA that codes for a luminescent protein, and thus cells that are infected by them will synthesize the protein. Since they express the spike protein on their surface, the pseudovirus initiates infection of cells in manner identical to SARS-CoV-2, thereby allowing researchers to study the mechanism of SARS-CoV-2 without having to handle a dangerous virus.
The pseudoviruses were incubated with a series of cell lines that expressed one of the 8 potential receptors on their own or in conjunction to ACE2. Cell lines that expressed one of the 8 candidate receptors but did not express ACE2 were not infected by the pseudovirus. However, 5 cell lines that expressed one of the candidate receptors in addition to ACE2 were infected at significantly higher rates than those that expressed ACE2 alone (Figure 4). This suggests that some of these receptors could be binding to the spike protein in conjunction to ACE2 to facilitate cellular infection. A similar experiment with pseudovirus expressing spike protein associated with the Delta strain of the virus was carried out. 7 out of 8 cell lines expressing one of the candidate receptors in conjunction to ACE2 were more susceptible to infection than those expressing ACE2 alone. This key and exciting finding suggests that the Delta strain of the virus is more transmissible in part due its ability to interact with more human receptors.
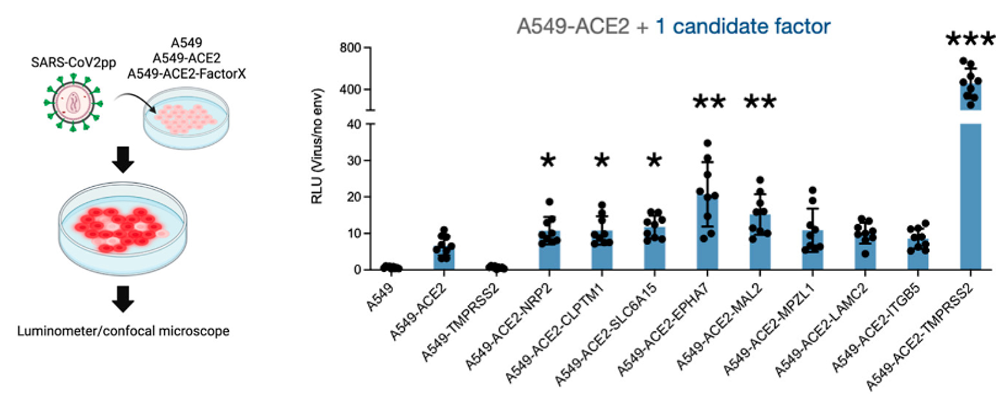
Figure 4 – (Left) Pseudovirus expressing spike protein is incubated with human cells. Infected cells express a luminescent protein that is encoded by the viral RNA. This allows researchers to quantify infection by the pseudovirus. (Right) Cell lines expressing ACE2 + one of the candidate factors are infected at higher rates than those expressing ACE2 alone, suggesting that the receptors also interact with the spike protein to facilitate infection by SARS-CoV-2. Reprinted with permission from J. Am. Chem. Soc. 2022, 144, 36, 16604–16611. Copyright 2022 American Chemical Society
In an experiment to understand the strength of binding between the receptors and spike proteins, the cells were incubated with a fluorescent molecule and the spike protein. Tracking changes in fluorescence levels could be used to measure how well the protein could bind to cells (Figure 5). Unexpectedly, there was no statistical difference in the levels of binding of the spike protein between cells that expressed ACE2 alone and those that expressed it in conjunction to one of the candidate receptors. This suggests that the interactions of these receptors to the spike protein may be transient, and so the receptors did not affect the amount of spike protein that was stably bound the surface of cells. However, more studies need to be carried out to determine how these receptors affect SARS-CoV-2 infection.

Figure 5 – (Top) Spike protein that has been tagged with a fluorescent molecule is incubated with one of the cell lines expressing the receptor proteins. Fluorescence levels are used to quantify the amount of Spike protein that has been able to bind to each of these cell lines. (Bottom) Cell lines expressing one of the receptors in addition to ACE2 bound the same levels of spike protein as the cell line expressing ACE2 alone. Reprinted with permission from J. Am. Chem. Soc. 2022, 144, 36, 16604–16611. Copyright 2022 American Chemical Society
Almost three years have passed since the discovery of COVID-19, but there are still aspects of SARS-CoV-2 infection that remain to be uncovered. This study illustrates the power of using proximity labeling techniques; identifying proteins that interact with each other can further our understanding of the mechanism of a viral disease that has changed the world in the last few years.