Featured image: PublicDomainPictures/Pixabay
Title: I-motif DNA structures are formed in the nuclei of human cells
Authors: M. Zeraati, D. B. Langley, P. Schofield, A. L. Moye, R. Rouet, W. E. Hughes, T. M. Bryan, M. E. Dinger D. Christ
Year: 2018
Journal: Nature chemistry 10.1038/s41557-018-0046-3
DNA is the molecule of life. It encodes all the information that determines how we look, how we function, how we grow and how we reproduce. For example, DNA determines the color of our eyes, the amount of hair, and the types of protein that we can express. Not only that, it also determines which species we are -if we are animal, if we are a bacteria, plant, or virus.
It is a polymeric molecule made of four nucleotides (Figure 1).
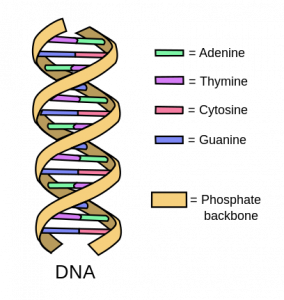
Figure 1: representation of double helix DNA and its main components
These nucleotides all have similar structures, with a sugar, a phosphate group, and a nucleobase (guanine, adenine, thymine, cytosine) (Figure 2). Since it only contains four types of nucleobases, we can think of DNA as a very long word written with only four letters (or a very long song written with only four notes). Different combinations of these nucleobases encode for all the proteins and all the information that is necessary for life to continue.
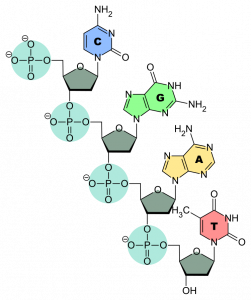
Figure 2: The structure of the nucleotides with the four nucleobases
The 4 nucleobases recognize each other in pairs of two – adenine + thymine and cytosine + guanine- and bind trough hydrogen bonds, forming the so-called base pairs. These bonds bring two complementary strands of DNA together to form double-stranded DNA (dsDNA, Figure 1) than then arranges in a double helix structure. These structures are usually supercoiled in the form of chromosomes that are located in the cellular nuclei (in the case of eukaryotes). The double helix can be unwound during some cellular phases – e.g. replication – by some regulatory proteins.
DNA can also assume other, more complicated structures like G quadruplex and i-motif (Figure 3), where the strands are unwound and separated in order to form some intra-strand hydrogen bonds.
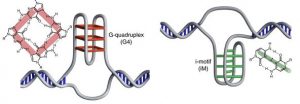
Figure 3: G- quadruplex and i-motif structures. Adapted with permission from I-motif DNA structures are formed in the nuclei of human cells, M. Zeraati et al., Nat. Chem. 2018. Copyright 2018 Springer Nature
The G quadruplex forms in regions that are rich in guanine content, where four guanine bases arrange in tetrads that stack in DNA helical motifs. G quadruplexes seem to have a role in the regulation of transcription of DNA and they are usually formed at the end of the DNA sequence of a chromosome, where there is an abundance of guanine bases.
I-motifs are similar structures, but they are a stack of intercalating base pairs formed between neutral and protonated cytosines, stabilized by acidic pH. They had been previously characterized in vitro, under acidic conditions -while the pH in the nuclei is slightly basic- but never directly seen in vivo, although the DNA sequence showed many regions rich in cytosine (because cytosine and guanine form a base pair, the strand that forms the i-motif is usually complementary to the one forming the G-quadruplex)
The authors of this paper synthesized an antibody able to recognize these i-motif structures with high affinity and were able to demonstrate their existence in the cellular environment for the first time.
After screening a library of deposited antibody fragments, they chose a segment that they called iMab and characterized its affinity and specificity for the i-motif in vitro under different conditions and in different positions on the DNA sequence.
At this point, they could move to the experiments in vivo where they use immunofluorescence staining to visualize the complex between iMAb and the DNA. With this technique, the antibody is labeled with a fluorescent probe: once iMab binds the i-motif, it can be visualized and localized within the cell nuclei thanks to its light emission. Further experiments showed that the concentration of i-motifs changes with the cell cycle, being maximum between the end of the cell growth and the beginning of DNA replication (G1/S boundary).
Indeed, every cell goes through a cycle of events that leads to the duplication of the cell (Figure 4). During this cycle, DNA has to be replicated so that the second cell can fully function. At the end of phase G1, before the replication begins (S phase), there is a checkpoint where the cell makes sure there are no mutations so to prevent the replication of dangerous cells (i. e. tumor cell). The higher concentration of i-motifs during this transition can imply a regulatory function in DNA replication.
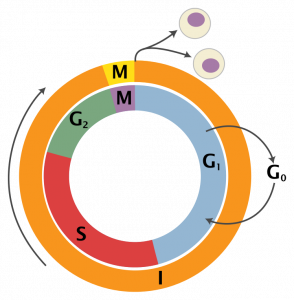
Figure 4: Schematic representation of cell cycle By Zephyris, derivative work: Beao and Histidine (talk)
Therefore, a mutation in the region where i-motifs are formed (e. g. the substitution of a cytosine with another nucleobase) could have implication in this step, and prevent it to fulfill its role.
A full understanding of its function in the cell cycle regulation could help move cancer therapy a step forward.
Thanks to Maria Nikolova for biology consulting.

