Featured Image: Image by Erin Rod / CC BY-SA 4.0, from Wikimedia Commons
Title: Machine-Learning-Guided Mutagenesis for Directed Evolution of Fluorescent Proteins
Authors: Y. Saito, M. Oikawa, H. Nakazawa, T. Niide, T. Kameda, K. Tsuda, and M. Umetsu
Journal: ACS Synthetic Biology
Year: 2018
https://dx.doi.org/10.1021/acssynbio.8b00155
Scientists studying living cells have a problem that at first sounds pretty simple and obvious: cells are tiny and hard to see! Even using microscopes, telling the difference between different parts of the cell is really difficult. Sometimes, we can use chemicals that give different colors to different cellular structures (called stains), but these often kill the cells they enter. However, there are plenty of biomolecules already inside living cells that don’t cause any problems, like proteins. What if there was a protein that glowed bright enough that a microscope could detect it? These exist and are called fluorescent proteins (Figure 1)!

Figure 1: Examples of fluorescent proteins in white light (top) and ultraviolet light (bottom). Adapted with permission using images (top, bottom) by Erin Rod / CC BY-SA 4.0, from Wikimedia Commons
Fluorescent proteins are part of a larger category of molecules that all exhibit a property called fluorescence. Basically, when illuminated with light of a certain color, these molecules emit light at a different color. The exact color of light absorbed and emitted depends on the chemical structure of the molecule. Some proteins have structures that can interact with light just like this! One of the most famous (and first discovered) is the simply named Green Fluorescent Protein (GFP), which glows green when ultraviolet light is shined on it.
Ever since GFP was first purified from glowing jellyfish, scientists have worked on discovering and even synthesizing entirely new types of fluorescent proteins, all with different characteristics. Proteins are made up of a string of smaller molecules called amino acids in a specific sequence. The exact arrangement of amino acids in a protein is what gives it its structure and function.
Usually, finding new fluorescent proteins is done by taking an existing protein and precisely switching individual amino acids with different ones and measuring the fluorescence properties of the new protein. But this is time consuming and there’s no guarantee that the new protein with be any better than the starting protein, or even be fluorescent at all. In this new paper, researchers have proposed a new way to use machine learning to carefully develop new and better fluorescent proteins.
These authors started with GFP and the goal of creating a new protein that emits yellow light. They started by doing the traditional method of fluorescent protein creation: modifying different parts of GFP and measuring both how bright the emitted light was (“relative maximum fluorescent intensity”) and how yellow it was (“yellow fluorescence ratio”). They compared these to both GFP and the already well-studied Yellow Fluorescent Protein (YFP), which they used as a reference. They made 218 different variants of GFP, but none of them were better than YFP (Figure 2).
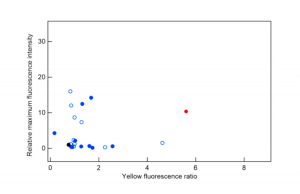
Figure 2: Fluorescent properties of randomly mutated variants of GFP (black circle) compared to reference YFP (red circle). Blue circles (solid and filled) are protein variants made using two different methods. Adapted with permission from Saito, Y.; Oikawa, M.; Nakazawa, H.; Niide, T.; Kameda, T.; Tsuda, K.; Umetsu, M. ACS Synth. Biol. 2018, 7(9), 2014-2022. Copyright 2018 American Chemical Society.
The authors took the 142 best performing proteins from this library, added GFP and YFP, and used these to train their machine learning program to look for proteins that had both high fluorescence intensity and yellow emission. Using this library, the program proposed new variant proteins that it determined had the highest chance of being high-performing, ranking them in that order. In an interesting outcome, the second-highest ranked variant protein actually had the same mutations as an already discovered version of YFP, called Venus. This showed that the predicted proteins were able to match the current methods of proposing new fluorescent proteins.
Of the proposed protein variants, the researchers synthesized and tested the top 78. All of them performed better than the starting GFP, with 12 of them having a higher yellow fluorescence ratio than even the reference YFP (Figure 3a). After they purified the proteins even further and measured the exact wavelength and intensity of their fluorescence emission, they found that the intensity decreased slightly (probably due to the difference in pH and solution components from the initial tests), but all of the proteins except one were even more yellow than YFP (Figure 3b).
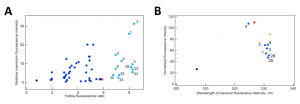
Figure 2: Fluorescent properties of randomly mutated variants of GFP (black circle) compared to reference YFP (red circle). Blue circles (solid and filled) are protein variants made using two different methods. Adapted with permission from Saito, Y.; Oikawa, M.; Nakazawa, H.; Niide, T.; Kameda, T.; Tsuda, K.; Umetsu, M. ACS Synth. Biol. 2018, 7(9), 2014-2022. Copyright 2018 American Chemical Society.
By studying these new structures of fluorescent proteins, the chemical link between molecular structure and fluorescent properties can be better understood. Scientists already know that changing certain amino acids influences specific aspects of fluorescence (such as intensity or emission wavelength), but machine learning lets them narrow their focus even more. Additionally, this way of finding new protein variants can be used to explore all kinds of different proteins, not just fluorescent ones. Changing what properties of the protein the program values can change the properties of the final variants.
Machine learning is becoming a valuable tool in many different fields for exploring huge amounts of data and proposing new, exciting solutions. With computers designing new versions of fluorescent proteins, biochemistry is no exception!

