Title: Quantification and Mapping of Alkylation in the Human Genome Reveal Single Nucleotide Resolution Precursors of Mutational Signatures
Authors: Yang Jiang, Cécile Mingard, Sabrina M. Huber, Vakil Takhaveev, Maureen McKeague, Seiichiro Kizaki, Mirjam Schneider, Nathalie Ziegler, Vera Hürlimann, Julia Hoeng, Nicolas Sierro, Nikolai V. Ivanov, and Shana J. Sturla
Journal: ACS Central Science
Year: 2023
DOI: https://doi.org/10.1021/acscentsci.2c01100
ACS Cent. Sci. 2023, 9, 3, 362–372
Chemical modifications to DNA can significantly impact cellular functions, affecting growth and contributing to disease development. A recent study explored DNA alkylation, a process where specific chemicals bind to DNA bases, causing damage that may result in mutations. The researchers sought to map these modifications at a single-nucleotide resolution, shedding light on the origins of mutational signatures within the genome.
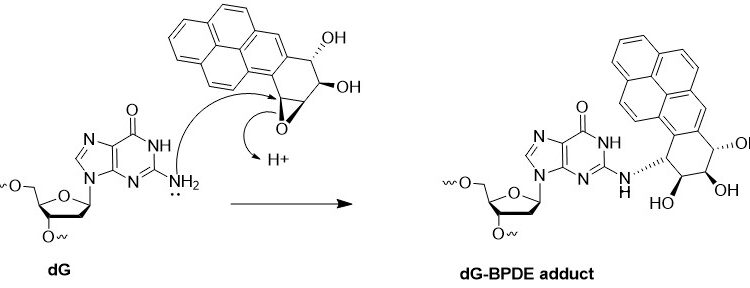
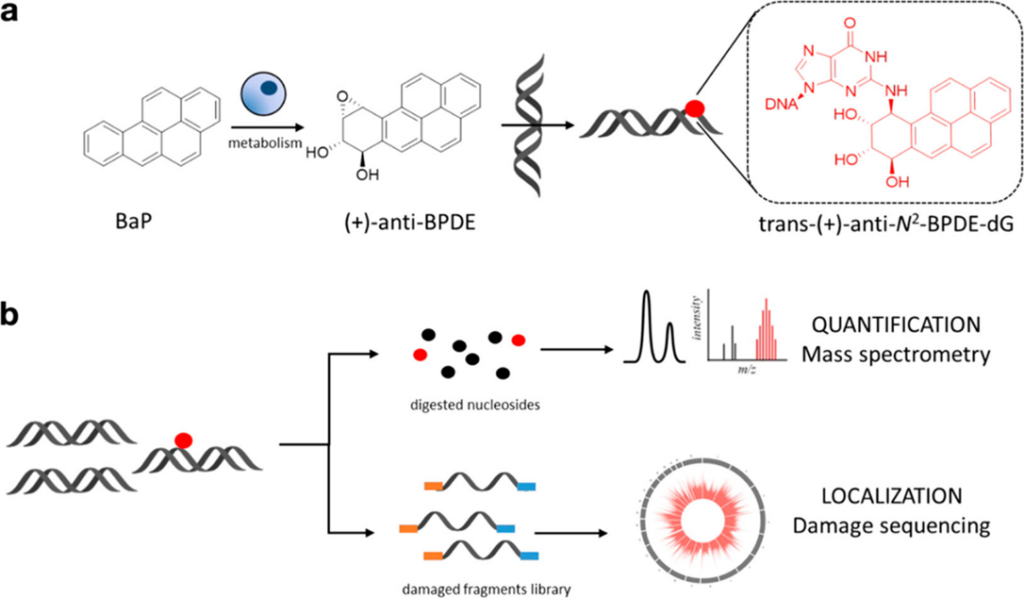
By developing a novel single-nucleotide resolution damage sequencing method, they mapped the main mutagenic adduct resulting from exposure to benzo[a]pyrene (abbreviated as B[a]P), a common environmental carcinogen. B[a]P is the primary representative of polycyclic aromatic hydrocarbons (PAHs), and has been repeatedly found in the air, surface water, soil, and sediments. It is present in cigarette smoke as well as in food products, especially when smoked and grilled. Human exposure to B[a]P is therefore common. B[a]P is metabolized into highly reactive compounds called B[a]P-diol-epoxides (BPDEs), which can react with DNA, forming DNA adducts, which are often harmful. The most abundant and mutagenic of these is N2-BPDE-deoxyguanosine (N2-BPDE-dG). DNA adducts like N2-BPDE-dG are typically repaired by nucleotide excision repair (NER). However, if they evade repair, they can lead to CG > AT mutations, commonly found in the cancer driver genes of smokers’ lung cancers. The mutational patterns observed in cells exposed to B[a]P closely resemble those found in smokers’ lung cancers.
Having precise DNA damage maps is crucial to better understanding the link between DNA adducts and mutational signatures. Traditional methods for mapping the distribution of N2-BPDE-dG have fallen short, lacking the single-nucleotide resolution needed to distinguish specific mutations like the CG > AT. Sturla and co-workers published a study in ACS Central Science that maps the locations of these DNA adducts at single-nucleotide resolution in the human genome. Their research reveals how these modifications correlate with mutations found in smoking-related lung cancers. The study employed two main techniques: quantification of N2-BPDE-dG levels and mapping its distribution in the genome. The researchers primarily used liquid chromatography-tandem mass spectrometry (LC-MS/MS). This method separates individual chemicals into a mixture and identifies them based on the mass and charge of their ions. To map the adduct locations, the researchers developed a method called BPDE-dG-Damage-seq. This method involves fragmenting genomic DNA, enriching for modified DNA through immunoprecipitation, and using a highly accurate DNA polymerase to identify adduct sites by stalling at these locations. You could think of the DNA polymerase as a car driving down the highway smoothly until it encounters a roadblock or a stop sign. Wherever this adduct exists on the genomic DNA, the DNA polymerase stops, allowing us to sequence these fragments and locate the exact positions of the adducts.
The results showed that DNA regions in open chromatin, which are more accessible, had fewer adducts compared to closed chromatin regions. Additionally, highly transcribed genes and their template strands were less damaged, indicating efficient repair mechanisms like nucleotide excision repair (NER) in these areas. Interestingly, the researchers observed that DNA damage accumulated significantly at the promoter regions of highly expressed genes but decreased sharply at the transcription start sites. This suggests that open chromatin regions are more prone to damage but are also more efficiently repaired. Interestingly, the distribution of DNA adducts correlated with known mutational signatures in lung cancer patients, particularly those associated with smoking-related cancers. They found consistent damage at specific mutation sites in essential cancer-related genes like CDKN2A (a critical tumor suppressor gene), SMARCA4 (the most frequently mutated ATPase in cancers), and many others! The observed correlation between B[a]P-induced DNA damage and mutations in these cancer-related genes underscores the carcinogenic potential of B[a]P and highlights its role in driving lung cancer development, especially in smokers.
This underscores the importance of reducing exposure to B[a]P-containing substances. The study further demonstrates the potential of using DNA damage maps to predict the mutational effects of chemical exposures. Future studies are expected to explore these patterns in different cell types over time to understand DNA damage and repair dynamics better.