Michelle Ehrenberger
Title: “Environmentally Benign Nanoantibiotics with a Built-in Deactivation Switch Response to Natural Habitats”
Authors: Zheng, Wan; Anzaldua, Miguel; Arora, Ankita; Jiang, Yunjiang; McIntyre, Kelly; Doerfert, Michael; Winter, Theodora; Mishra, Abhijit; Ma, Hairong; Liang, Hongjun.
Journal: Biomacromolecules
Year: 2020
https://pubs.acs.org/doi/10.1021/acs.biomac.0c00163
Featured image, figures, and table reproduced with permission. Further permissions related to this material must be directed to the ACS.
In 2010, 90,000-180,000 tons of antibiotics were released into the environment as a byproduct of antibiotic use in human healthcare and agriculture. However, many strains of bacteria compose a vital part of natural ecosystems, breaking down organic matter such as fallen leaves and building new nutrients that can be utilized by plants and animals. The massive amounts of antibiotics released into soil and water can upset this system of nutrient recycling, destabilizing ecosystems while threatening food and water security for humanity. Continual exposure to antibiotics in the environment can also put selective pressure on bacteria to develop resistance genes against them. Because bacteria can transfer DNA between strains and species, a resistance gene originating in a non-pathogenic bacterial strain could be transferred to a pathogenic strain, eventually leading to an increase in multidrug-resistant infections. Because of this, new strategies must be developed to preserve the lifesaving benefits of antibiotic use in medicine while eliminating this environmental contamination.
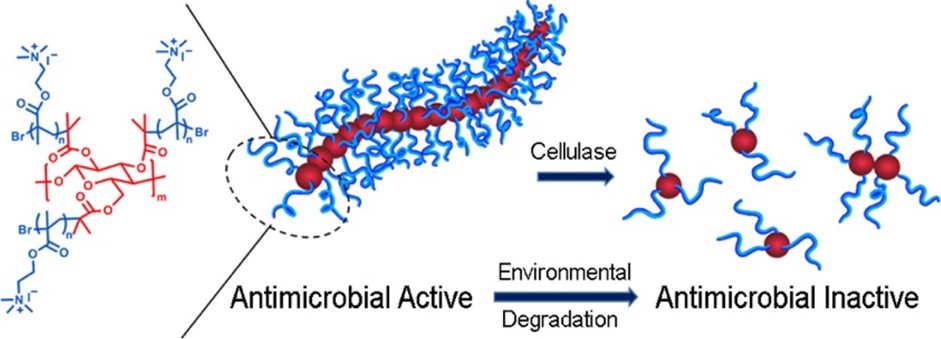
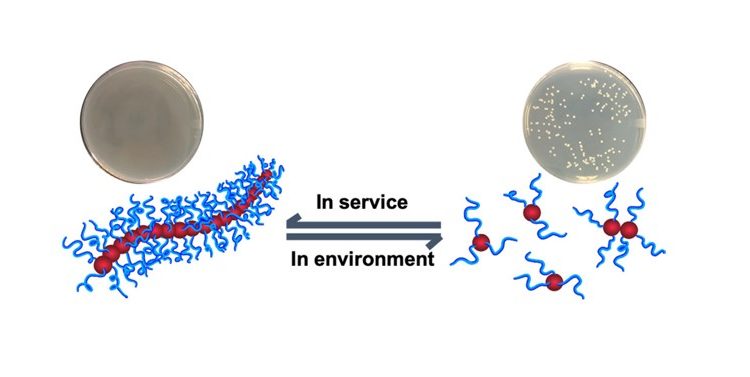
To solve this problem, Zheng and co-workers have designed an antibiotic nanoparticle that is active in the human body but is naturally degraded when released into the environment. The nanoparticle, called a Polymer Molecular Brush (PMB), consists of a cellulose backbone covered in hydrophilic (water-soluble), linear “brush” polymers (Fig. 1). The PMBs were able to kill both Gram-positive and Gram-negative bacteria, including multidrug-resistant strains, but not human red blood cells (HRBC, Table 1) or kidney cells.
Table 1. Biological effects of PMBs, cellulase-degraded PMBs, and linear brush polymers (called PTMAEMA32). MBC = minimum bactericidal concentration. MIC = bacterial inhibitory concentration. HC50 = hemolytic cytotoxicity, the concentration required to kill 50% of red blood cells. E. coli and PA14 are Gram-positive. S. aureus and MU50 are Gram-negative. Both PA14 and MU50 are multidrug-resistant strains.

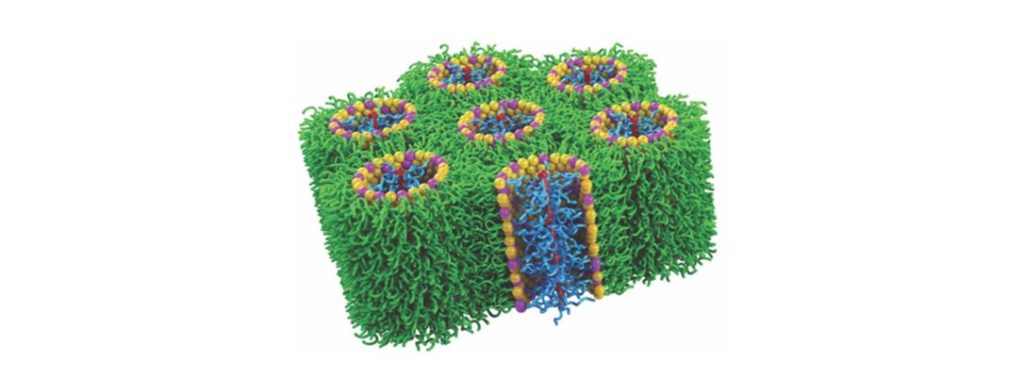
The PMB antibiotic activity comes from its ability to form holes in bacterial membranes, destroying the barrier between inside the cell and outside. As can be seen in a computer model (Fig. 2), the cellulose backbone lies along the center of the newly formed pore. The hydrophilic brush polymers interact with the hydrophilic heads of the membrane lipids, while the hydrophobic (non-water soluble) tails of the membrane lipids are excluded. The bacteria are killed because they can no longer regulate what molecules—including water—are allowed to enter or leave through the membrane. Human cell membranes are not affected by PMBs because of the difference in lipid composition between human and bacterial membranes. Bacterial membranes naturally have regions of negative curvature, like craters. This inherent curvature means it takes less energy for the PMBs to rearrange bacterial membranes to form these pores. Human cell membranes lack this curvature, so PMBs are unable to rearrange the lipids or kill human cells.
The environmental degradation of PMBs, however, stems from the use of cellulose as their backbone. Cellulose is the natural building material used to give structure to plant cell walls, for example, the strong fibers in a celery stick. Cellulases are enzymes that break the polymer down into its individual building blocks (Fig. 1). Importantly, cellulases are found commonly in the environment—made by bacteria, protozoa, and fungi that can break down plant material—but are not found in the human body! The scientists found that their PMBs could be broken down by cellulases in only 3 hours, and that these degraded PMBs were no longer active against bacteria (Table 1). Thus, this nanoparticle antibiotic should be functional in humans but lose function after being released into the environment.
Antibiotic medicine is often the deciding factor in whether an infection is inconvenient or deadly. However, the current unchecked release of antibiotics into the environment can not only decrease the effectiveness of antibiotics over time through the development of multi-drug resistance but can also destabilize naturally balanced ecosystems in unpredictable ways. The PMB nanoparticle antibiotic design offers one solution to this problem. More research is still needed before PMB nanoparticles can be used in clinical trials on humans. For example, when linear brush polymers alone (without their cellulose backbone) were introduced to human red blood cells, the red cells clumped together in a process called hemagglutination, but the polymer’s role in this process is not yet known. Despite this, PMB nanoparticle design is a new and innovative strategy toward making antibiotic medicine more sustainable.