Primary source information:
Title: Identifying molecules as biosignatures with assembly theory and mass spectrometry
Authors: Stuart M. Marshall, Cole Mathis, Emma Carrick, Graham Keenan, Geoffrey J. T. Cooper,
Heather Graham, Matthew Craven, Piotr S. Gromski, Douglas G. Moore, Sara. I. Walker & Leroy Cronin
Journal: Nat. Commun (2021) 12:3033
Earlier this year, NASA’s Perseverance rover landed on Mars with the stated goal to seek signs of life. If there has ever been microbial life on Mars, or elsewhere in the solar system, how will we detect these organisms? In order to detect signs of life (called biosignatures), we must simultaneously confirm that the biosignature can be produced by living organisms, and rule out the possibility that the biosignature can be produced in the absence of organisms. Organisms on Earth have been well characterized, so we know that biomolecules like DNA and phospholipids provide unambiguous signatures of life. However, alien biochemistry might be unimaginably different than our own, so the task of detecting alien life is particularly challenging.
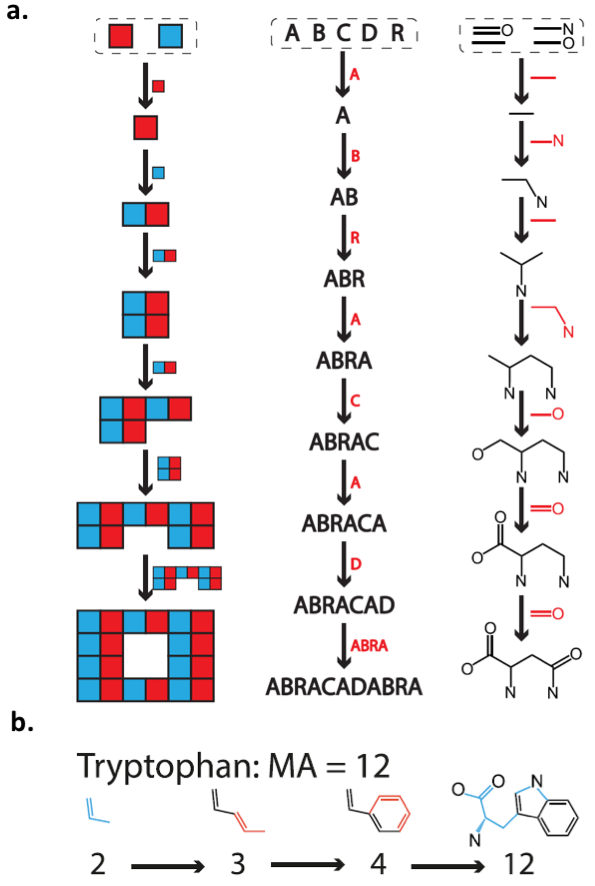
In this paper by Marshall et al., the authors suggest that all living organisms will produce some kind of complex molecule that can not be produced in the absence of life. The authors describe the “complexity” of a molecule as relating to the molecule’s size and symmetry, and they quantify this complexity using a new framework called assembly theory (Figure 1). By starting with a set of chemical bonds, the authors enumerate all the unique ways to assemble a given molecule from those bonds. The simplest way to assemble the molecule from its component atoms and bonds is called the molecular assembly number (MA). Molecules with very high MA are unlikely to form in the absence of life, and thus can be used as biosignatures. It is important to note that this is a conceptual tool: the assembly steps do not necessarily reflect real chemical reactions.
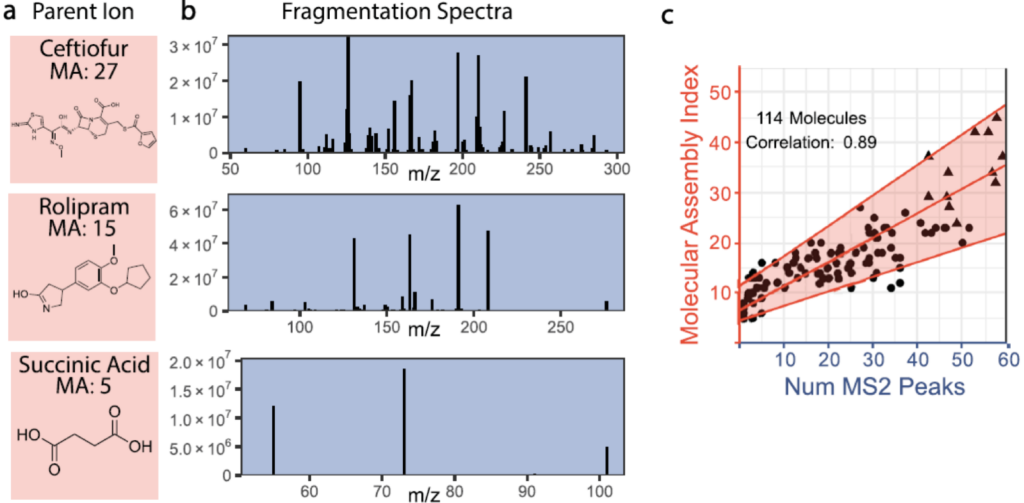
Next the authors show that for an unknown molecule, they can accurately estimate the molecular assembly number using tandem mass spectrometry (MS2). In tandem mass spectrometry, molecules are broken apart into fragments by collisions with an inert gas, then the fragments are separated based on their mass-to-charge ratio. The number of peaks in the mass spectrum corresponds to the number of unique fragments that were generated from the original molecule. As a result, larger molecules with less symmetry tend to produce more unique fragments than smaller, symmetric molecules. The authors test over 100 different molecules, and find that the number of peaks in the mass spectrum is a good predictor of the molecular assembly number (Figure 2).
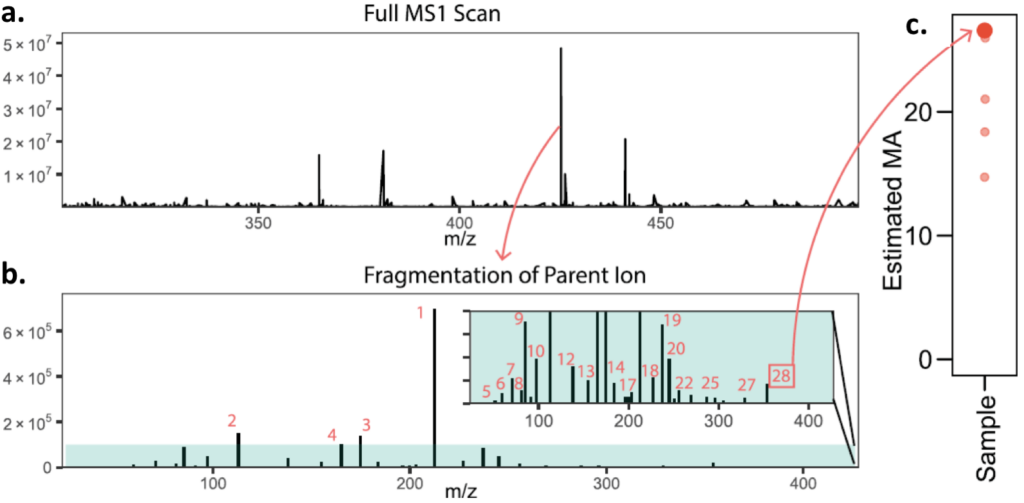
The authors also test their approach on an unknown mixture of molecules. First, the authors use mass spectrometry to separate all of the molecules based on their mass:charge ratio (m/z). This initial mass spectrum is called MS1 (Figure 3A). Then the authors isolate one of the molecules (parent ion) and generate fragments (Figure 3B). They repeat this procedure for each parent ion, and count the number of fragments in each case. The maximum number of fragments corresponds to the molecular assembly number for the original mixture (Figure 3C). This approach is ineffective when two parent molecules have the same mass:charge ratio. In this case, the apparent number of fragments will be greater than the number of fragments from each individual parent ion, leading to an overestimate for the molecular assembly number. However, there are strategies to mitigate this problem. First, by using high resolution mass spectrometry, the authors can reduce the likelihood that two parent molecules will have the same mass:charge ratio. Second, the authors could use chromatography to separate the molecules based on physical properties (e.g. hydrophobicity) prior to mass spectrometry.
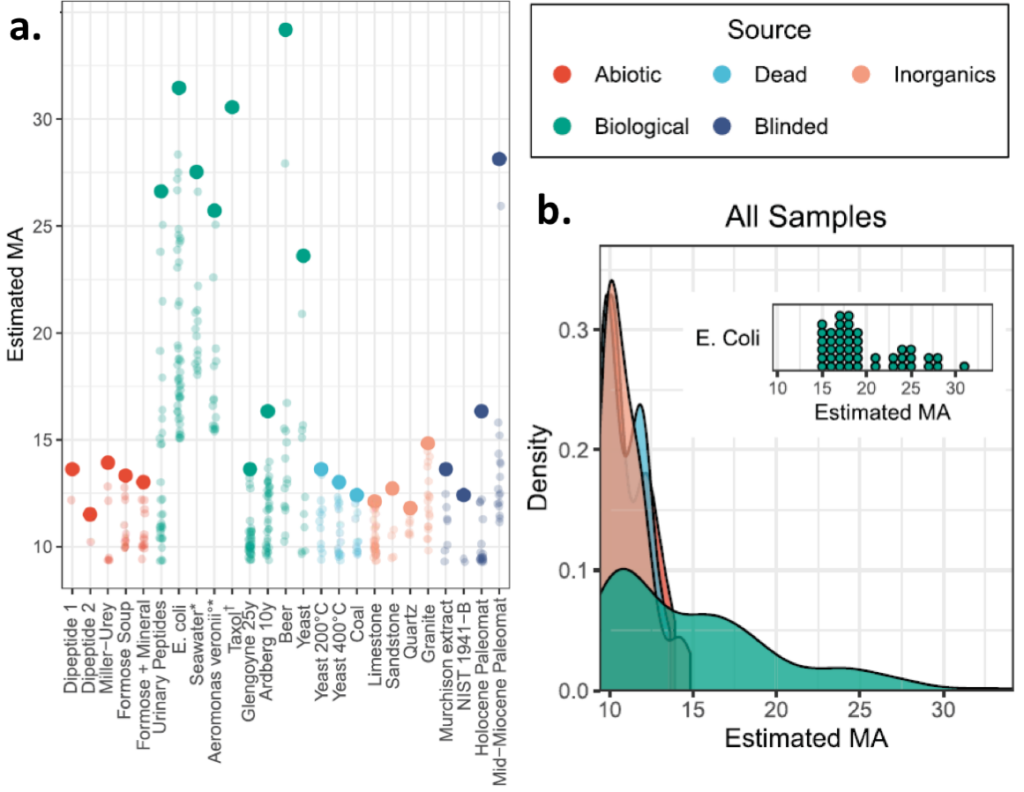
The authors apply their approach to a number of unknown mixtures from various sources. For example, they compare biological samples (including cell lysates and ecological samples) with inorganic minerals and non-living chemical reaction products. Some of the non-living chemical reactions are particularly interesting because they represent plausible steps in the origin of life (e.g. Miller-Urey and Formose). In all cases, the authors are able to distinguish the biological samples from non-biological samples (Figure 4). These findings validate the authors claim that large molecular assembly numbers can be used as biosignatures! This is a promising development for the detection of alien biosignatures from Mars sample return missions!