Article Title: Stereoselective biocatalytic α-deuteration of l-amino acids by a pyridoxal 5′-phosphate-dependent mannich cyclase
Authors: Gao, J.; Zhou, C.; Hai, Y.
Journal: ChemBioChem
Year: 2023
DOI: /10.1002/cbic.202300561
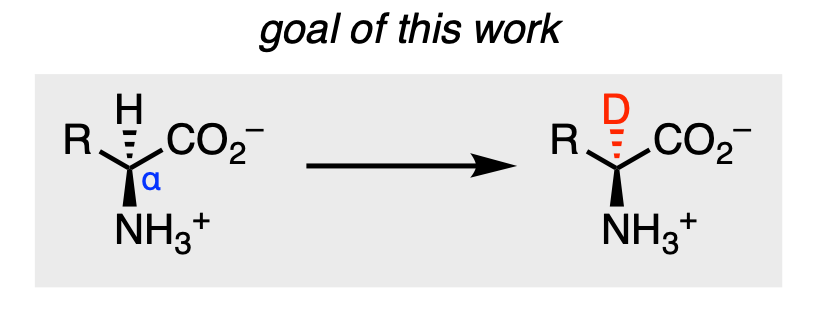
Hydrogen is the most abundant element in the universe and is part of nearly every known compound, including drugs. Given its prevalence, there is general interest in developing methods to substitute hydrogen (H) for its isotope deuterium (D) within compounds. Deuterium contains the same number of protons and electrons as hydrogen but is heavier by one neutron. Such changes can alter properties of the original compound, such as its acidity, without greatly changing the molecule’s overall function. Most importantly, the stability of drugs can often be improved as deuterium is less reactive. The carbon–deuterium (C–D) bond is stronger than the carbon–hydrogen (C–H) bond, which makes it more difficult to break apart a molecule at that site. This principle is known as the kinetic isotope effect. Given this knowledge, Gao and coworkers set out to engineer enzymes to deuterate (add deuterium) amino acids at the α‑position (Figure 1).
Chemists can develop organic synthesis methods to add deuterium atoms; however, these typically require multiple steps if there are multiple places where a deuterium atom could go in a target molecule. Unlike typical organic reactions, enzymes can catalyze reactions at specific locations within molecules and produce only one, rather than two, possible isomer products. This ability is known as a regio– and stereo–selectivity. Gao et al. decided to target the enzyme LolT, as they had previously worked with this enzyme. LolT is involved in producing the insecticide loline (Figure 2A). Based on previous work, they knew that LolT is a pyridoxyl phosphate (PLP)-dependent enzyme that catalyzes an intramolecular cyclization reaction—in other words, a ring is formed within the molecule itself.
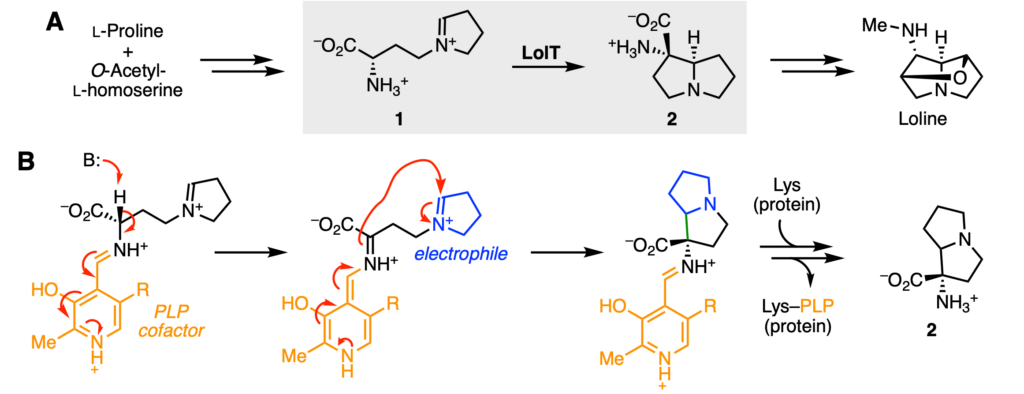
The proposed mechanism of LolT is similar to ones proposed for other PLP-dependent enzymes (Figure 2B). In brief, the cofactor PLP (orange) is reused by the enzyme and serves as an “electron sink” to promote the chemical reaction. After the molecule binds, a proton (H+) is removed, and the electrons are shuttled down to the positively charged amine (NH+) of PLP. Next, the electrons are shuttled back up and attack the electrophilic (“loves electrons”), positively charged imine (C=N+, blue) of the molecule. As a consequence, a new carbon–carbon bond (green) is formed, resulting in a new cycle.
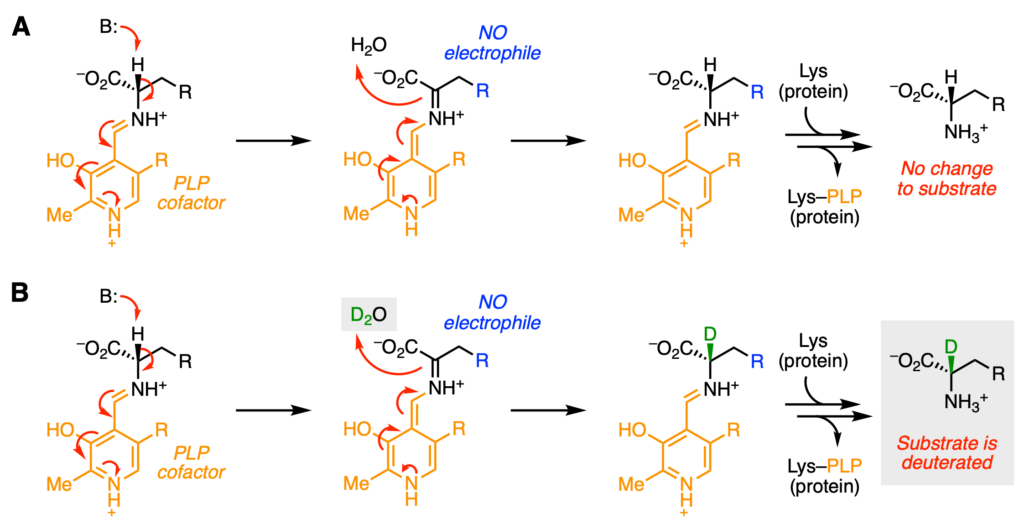
The reactivity of LolT can be changed by altering the molecule identity. For instance, in the case a molecule without an electrophilic group is used (Figure 3), LolT will become stuck. The first step deprotonation step can proceed as normal. However, in the second step, there is no electrophile in the molecule for the electrons to go into. Instead, the electrons will grab a proton from the most abundant chemical—the solvent, or H2O. As a result, the molecule is unchanged after the enzyme reaction (Figure 3A). However, if the reaction is run in heavy water, or D2O, the electrons will grab a deuteron instead! Thus, a new C–D bond is formed (green) and the molecule is deuterated (Figure 3B).
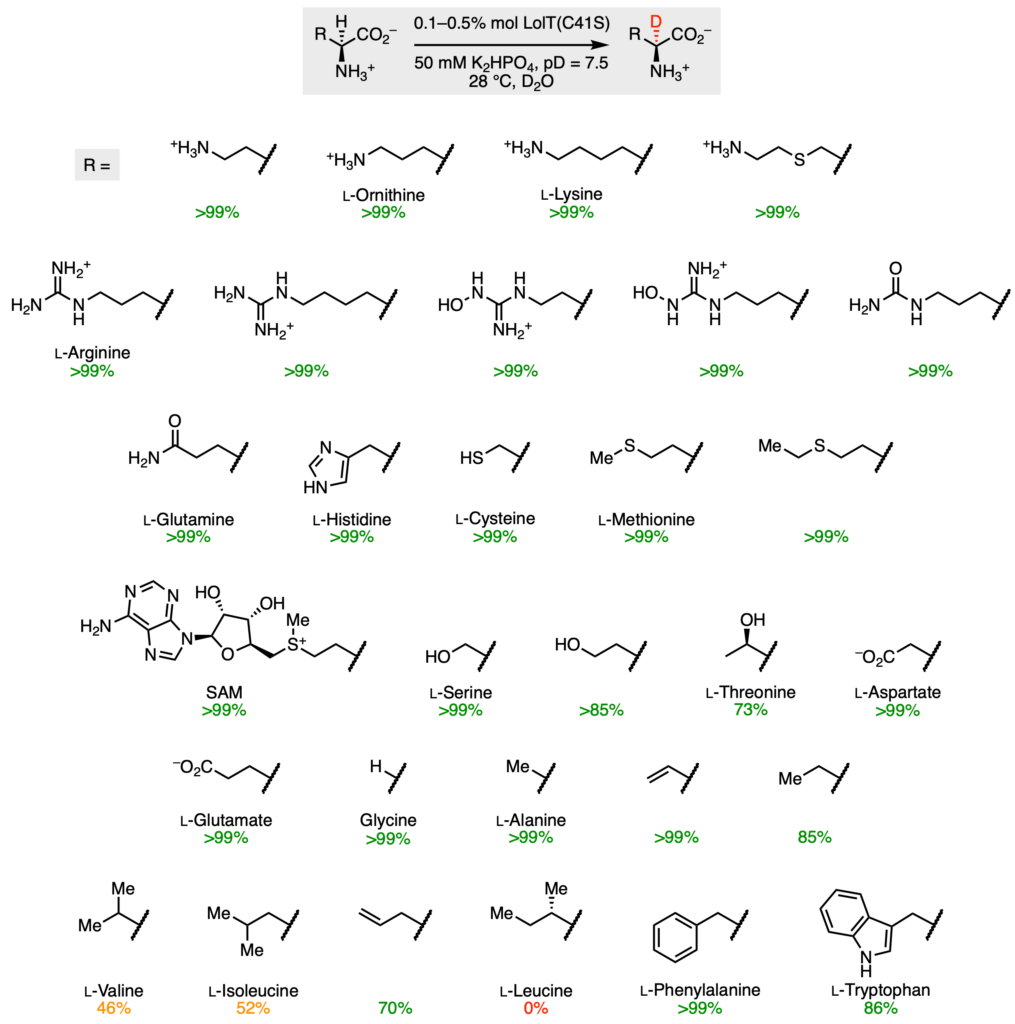
Using this principle, Gao and coworkers tested a variety of alternative molecules to figure out the natural substrate scope (Figure 4). Ideally, a single enzyme can be used to broadly add deuterium atoms to multiple different molecules. LolT selectively deuterated multiple enzymes with high efficiency, including most of the common amino acids that make up proteins in our body. The exceptions were valine, isoleucine, and leucine; all branched molecules. One possible explanation for this result is that these molecules are too bulky to fit within the enzyme active site. In addition, other very common molecules found within our bodies, such as ornithine and SAM, could also be deuterated at the α‑position.
In this work, the PLP-dependent enzyme LolT was used to deuterate the α‑position of multiple different amino acids. By understanding the mechanism of how LolT catalyzes its natural reaction, scientists were able to manipulate the reactivity of LolT. Now, scientists can access many deuterated molecules that can be used to make deuterated, more stable drugs. Although this is not the first example of using PLP enzymes to deuterate molecules, LolT has an exceptionally broad substrate scope compared to other PLP enzymes, making it of high utility. Continual study of enzyme mechanisms more broadly will also further our ability to develop novel chemical reactions by similarly tweaking the reaction conditions.

