Title: Spatial imaging of glycoRNA in single cells with ARPLA.
Authors: Yuan Ma, Weijie Guo, Quanbing Mou, Xiangli Shao, Mingkuan Lyu, Valeria Garcia, Linggen Kong, Whitney Lewis, Carson Ward, Zhenglin Yang, Xingxin Pan, S.Stephen Yi & Yi Lu.
Journal Nature Biotechnology
Year: 2023
doi: 10.1038/s41587-023-01801-z
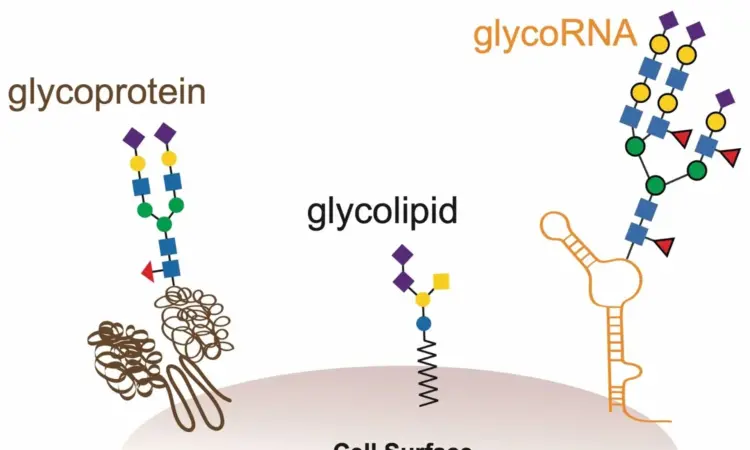
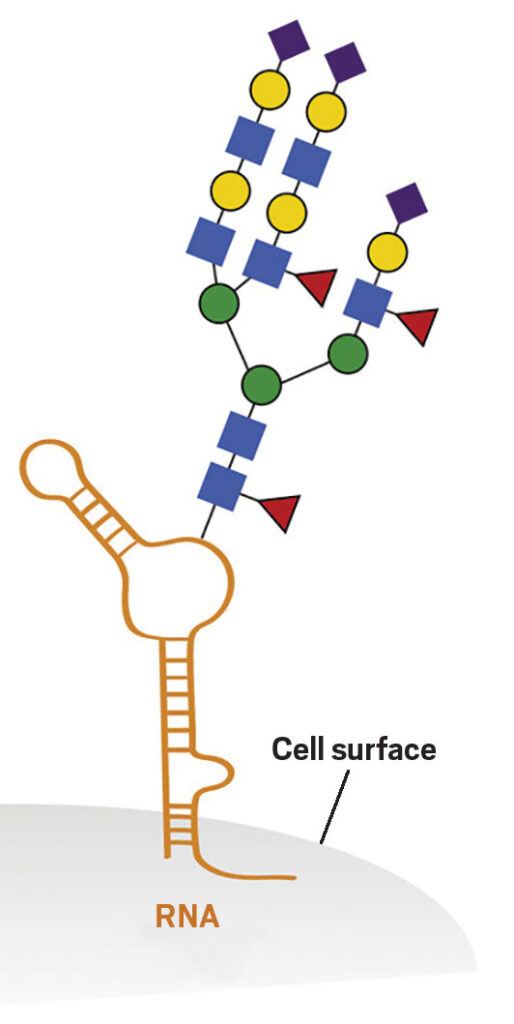
Cellular glycans are polysaccharides, which are giant biomolecules with multiple connected sugars. They are crucial for regulating essential biological functions, including cell communication, immune modulation, and embryonic development. Recently, glycosylated RNAs (glycoRNAs) have been discovered on various cell surfaces, sparking interest due to their potential roles in RNA-related processes. GlycoRNAs are small, non-coding RNAs that have been modified with sugar molecules. These RNAs are coated with a class of sugars known as N-glycans. The discovery of these biomolecules by Ryan Flynn, MD, Ph.D. at Stanford University, in the lab of Carolyn Bertozzi, Ph.D., revealed that some of the glycoRNAs’ N-glycans were capped by a sugar called sialic acid. GlycoRNAs are an absolute game-changer in the realm of cancer biology! They hold immense potential to revolutionize our understanding of cancer progression by unveiling the secrets behind cancer cell migration, invasion, and drug resistance. The discovery that aberrant glycosylation patterns underlie the very essence of cancer’s devious nature is ground-breaking because there was no previously known intersection between the RNA world and the glycan world.
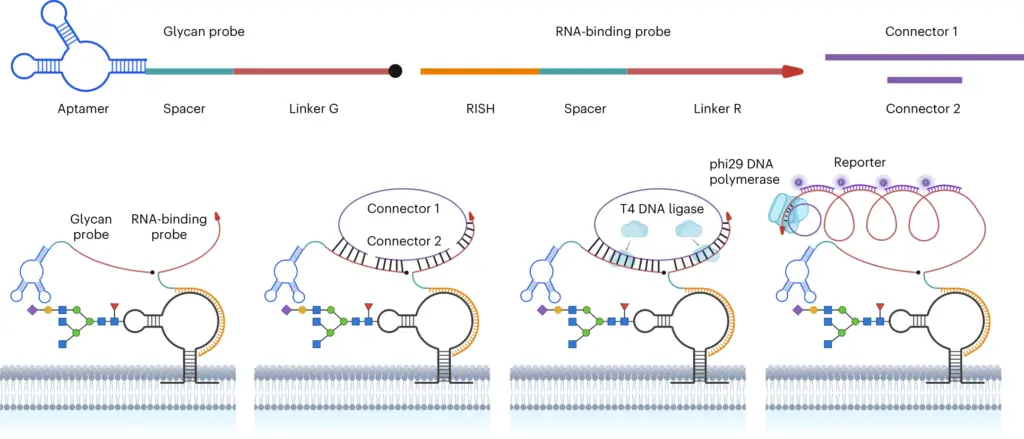
Figure 1. General working scheme for ARPLA
Scientists have long employed various methods to study glycoRNAs. The researchers first used metabolic labeling and click chemistry-based RNA blots to confirm the existence of glycoRNAs. Then, next-generation sequencing was used to profile what RNA sequences are found in glycoRNA. High-performance liquid chromatography-mass spectrometry was used to identify the composition of sugars in the glycans. Finally, and most recently, antibodies, or glycan-binding proteins, verified the presence of glycoRNA on cell surfaces. Despite these advances, none provide both sequence and spatial information, a critical connection required to understand the properties and functions of glycoRNAs better.
To address this challenge, Ma et al. recently invented a spatial imaging technique named the sialic acid aptamer and RNA in situ hybridization-mediated proximity ligation assay or ARPLA and successfully visualized glycoRNAs in single cells for the first time. Let us break down this technique a little bit; aptamers are DNA sequences that can selectively bind to a particular analyte. This analyte can be a small molecule or a protein, and in this case, the aptamer recognizes sialic acid selectively! A complementary DNA probe is used for RNA recognition by hybridization within the cell. By dual recognition of the sialic acid and the RNA probe, when these two probes come close to one another in a cell, they can become connected and form a circular piece of DNA. A process called rolling circle amplification (RCA) then amplifies this circular DNA, enabling a third probe to detect it. This third DNA probe is labeled with a fluorophore so that when it binds to the amplified DNA, it generates a fluorescent signal. These fluorescent signals are robust and distinct, offering high sensitivity for studying the spatial distribution of glycoRNAs. By substituting the specific sequence of the DNA probe, different RNAs, and thus glycoRNAs, can be identified. This incredibly versatile tool has the potential for visualizing a wide range of glycoRNAs.
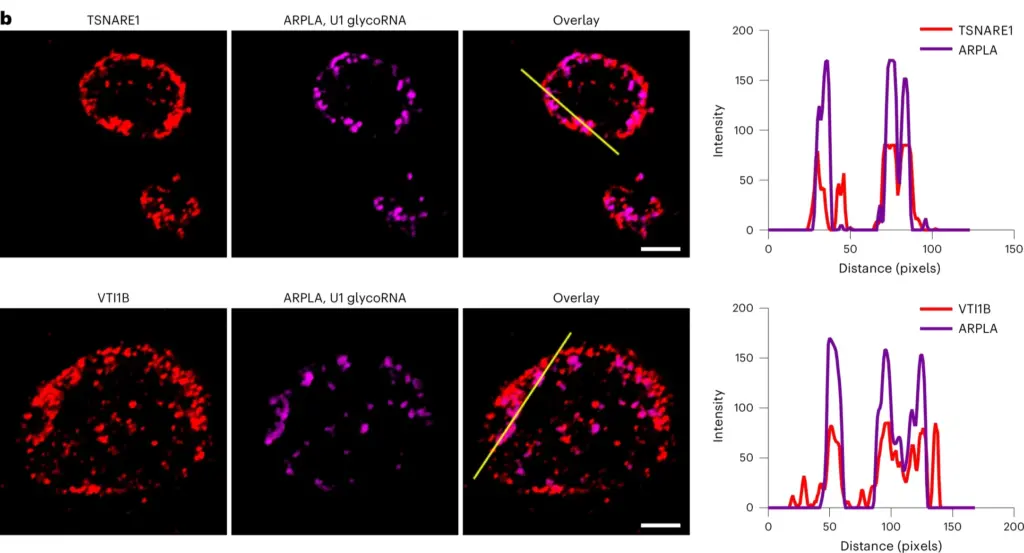
Figure 2. Fluorescence imaging of SNARE proteins indicated by Alexa Fluor 555 (Red) and its colocalization with glycoRNAs imaged by ARPLA
This method enables scientists to investigate the interactions between glycoRNA and specific regions on the outer surface of cells known as lipid rafts. Using ARPLA, the researchers could map where glycoRNAs are distributed on cell surfaces and found that they tend to cluster around lipid rafts. This finding has significant implications because it suggests that glycoRNAs may directly influence cell-to-cell communication, a role that was not well-established previously. The mechanism by which glycoRNAs are transported within cells to reach the cell surface could also be identified. They are guided by proteins called SNARE proteins that help release cell contents.
The ARPLA technique is now being applied in the field of breast cancer research. This approach provides visual insights that suggest an interesting pattern regarding the levels of sugar-modified RNA molecules (glycoRNAs) on the cell surface and their association with tumor aggressiveness and the likelihood of cancer spreading, which is known as metastasis. It offers a new way to predict the progression of the disease. For instance, in this research, they found that non-cancerous breast cells had the highest levels of glycoRNAs on their surface, followed by breast cancer cells with varying levels, and the metastatic breast cancer cells had the lowest levels. This implies that a higher concentration of surface glycoRNA is linked to a lower probability of cancer spreading to other parts of the body. Investigating the role of glycoRNAs in immune cell dynamics revealed that as human leukemia monocytic cells transformed into macrophages, the levels of glycoRNA decreased. This observation opens up promising research opportunities to understand how glycoRNAs impact immune cell functions.
Visualizing glycoRNAs has the potential to unveil their hidden roles in cancer biology, disease progression, and immune cell studies, promising a treasure trove of undiscovered insights. ARPLA not only serves as the inaugural cornerstone in elucidating the intricate spatiotemporal dynamics of this elusive biomolecule but also boasts adaptability. Through a judicious exchange of the glycoRNA recognition probes with counterparts tailored for various biomolecules, including modified RNAs, modified DNAs, and glycoproteins, ARPLA emerges as a versatile conduit for the systematic exploration of diverse molecular domains.