Title: A Plastic Biosynthetic Pathway for the Production of Structurally Distinct Microbial Sunscreens
Authors: Sıla Arsın, Endrews Delbaje, Jouni Jokela, Matti Wahlsten, Zoë M. Farrar, Perttu Permi, and David Fewer
Publication: Chemical Biology
Year: 2023
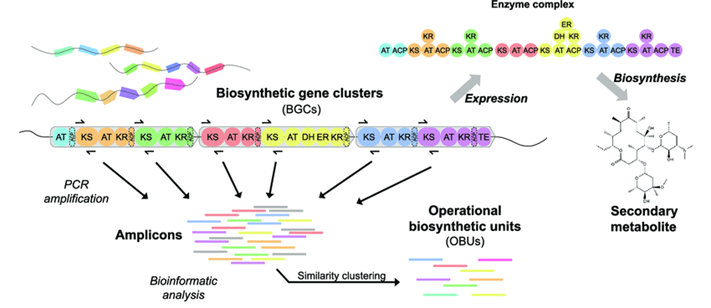
Sunscreen protects your skin from harmful ultraviolet (UV) rays through two methods—acting as a physical shield to reflect UV rays or absorbing UV rays. Recently, a class of molecules called “mycosporine like amino acids (MAA)” were shown to absorb UV rays and possess antioxidant properties. There are about 70 variants of these molecules, and they are metabolites produced by a bacterial genus called Nostoc. Structurally, they contain a cyclohexanimine chromophore, with substituents can be an amino acid and/or sugar moiety. A chromophore is a group that contains conjugated double bonds which usually imparts color to the compound. (The cyclohexanimine is highlighted in red Fig.1 A). Bacteria produce, or “biosynthesize” these molecules using enzymes encoded next to one another within the genome (shown in Fig 1 B). These “clusters” of enzymes are called “biosynthetic gene clusters”.
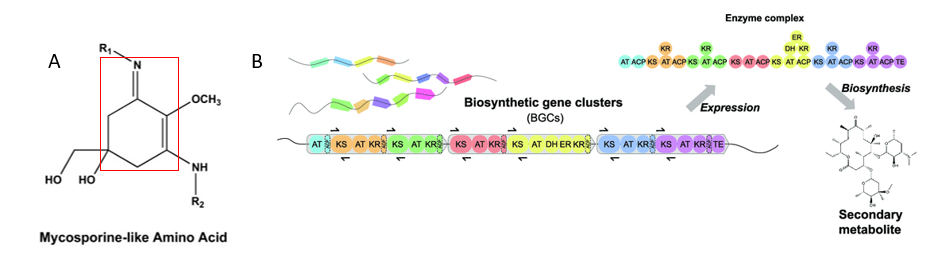
Sula et al. identified two MAA variants in the species Nostoc sp. UHCC 0926. These two novel structures look chemically dissimilar from known MAAs, indicating that they might be encoded by different biosynthetic gene clusters. The MAAs are named Tricore B and Aplysiapalythine E (Fig. 2). The scientists used two major tools to characterize these structures—ultra-performance liquid chromatography with quadrupole time of flight (UPLC-QTOF) and nuclear magnetic resonance (NMR) spectroscopy. UPLC-QTOF uses liquid chromatography to separate compounds based on hydrophobicity in tandem with mass spectrometry to accurately determine the mass-to-charge ratio of compounds. NMR uses magnetic fields to detect specific nuclei in the compounds, providing insight into the atom connectivity and thus, structure.
Due to the structural dissimilarity between Tricore B and Aplysiapalythine E, the authors reasoned that these compounds used distinct biosynthetic pathways as well. To test this, they sequenced the genome of Nostoc sp. UHCC 0926 and identified three potential distinct clusters as shown in Fig 2. The first gene cluster contained the genes mysD, mysC2 and mysC3; the second contained mysA, mysB and mysC1; and the third had mysF, mysI, and mysE. Upstream of mysF, the authors found a mysC1 like gene, however, it doesn’t code for the corresponding protein. Two of these clusters were located on the chromosome while the third was on a plasmid. Plasmids are circular DNA molecules that are different from the cell’s chromosome. Based on the predicted functions of these genes, the authors proposed the biosynthesis of Tricore B and Aplysiapalythine E depicted in the scheme of Fig 3.
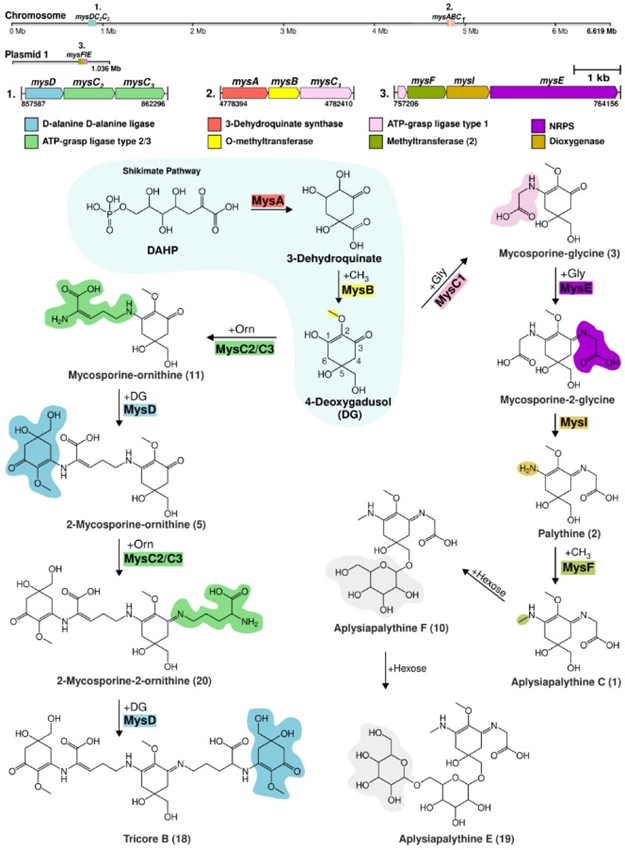
This scheme was pieced together using chemical logic. For instance, to make the left, center, and right side moiety (blue) of Tricore B, the authors realized that the most likely precursor molecule is DAHP from the pentose phosphate pathway. MysA, a predicted cyclase enzyme, converts DAHP into the 6-membered ring form, 3-dehydroquinate. MysB, a predicted O-methyltransferase, likely catalyzes the addition of a methyl group to the oxygen of 3-dehydroquinate to form DG (blue). To extend the structure, either MysC2 or MysC3, both predicted ATP-grasp ligases, adds the amino acid ornithine (green). A second molecule of DG is added by MysD, another ligase. These final two steps are repeated once more to yield Tricore B. A similar biosynthetic pathway was proposed to produce Aplysainpalythine E as shown in the figure above. These pathways though are hypothetical, and experiments need to be done to validate these.
Having identified a unique configuration, the scientists were curious if such discontiguous biosynthetic pathways were common in other bacteria. They compared the genomes of 75 bacteria that contained biosynthetic gene clusters for MAA biosynthesis. Of these, 9 contained the same discontiguous MAA architecture like Nostoc sp. UHCC 0926. The authors generated a phylogenetic tree of the bacteria to explore how these clusters may have evolved. By comparing key genes shared by all 75 bacteria, the authors could make a “bacterial family tree.” Bacteria with very dissimilar genes likely evolved separately from one another further in the past than bacteria with very similar genes. Since the 9 bacteria with discontiguous MAA biosynthetic gene clusters were found in multiple, separate places in the phylogenetic tree, they hypothesized that a process called horizontal gene transfer could explain the gene cluster’s origin. Horizontal gene transfers occur among bacteria and viruses where genetic information can cross barriers and transfer from one bacterium to another through various mechanisms.
The results of this study are promising. It indicates that through various mechanisms, bacteria have evolved to produce compounds to protect themselves. The authors used a type of cyanobacteria and unlike most bacteria, they use light to make energy, so they are exposed to sunlight and hence need to produce molecules to protect themselves. Further, because this study shows that discontiguous gene clusters can be coordinated to produce a new molecule, there is a potential for more diverse molecules to be biosynthesized as discontiguous clusters work in tandem with each other. More studies are yet to be done to confirm this and explore the full potential of this discovery. Besides this, identification of novel biosynthetic gene clusters has the potential to discover new compounds with beneficial properties that can be harnessed as drugs.
