Title: Photoactivatable Glycolipid Probes for Identifying Mycolate–Protein Interactions in Live Mycobacteria
Authors: Benjamin M. Swarts, et al.
Journal: JACS
Year: 2020
https://pubs.acs.org/doi/10.1021/jacs.0c01065
A mycobacterium is quite the hardy microbe! Organisms in this genus arm themselves with a hard-to-penetrate armor made up of not just the standard peptidoglycan wall, but also a waxier, lipid-rich coating known as the mycomembrane. This mycomembrane is weaved from a dense array of long fatty acids (mycolic acids) linked via ester bonds to sugars, which in turn connect the entire structure to the canonical peptidoglycan wall (Fig. 1). This seal-tight assembly contributes to these microbes’ hardiness and makes them nefarious pathogens that are notoriously hard to treat. The most infamous species, Mycobacterium tuberculosis (which causes tuberculosis) and Mycobacterium leprae (which causes leprosy), still plague our healthcare systems and pose burdens to millions of lives yearly.
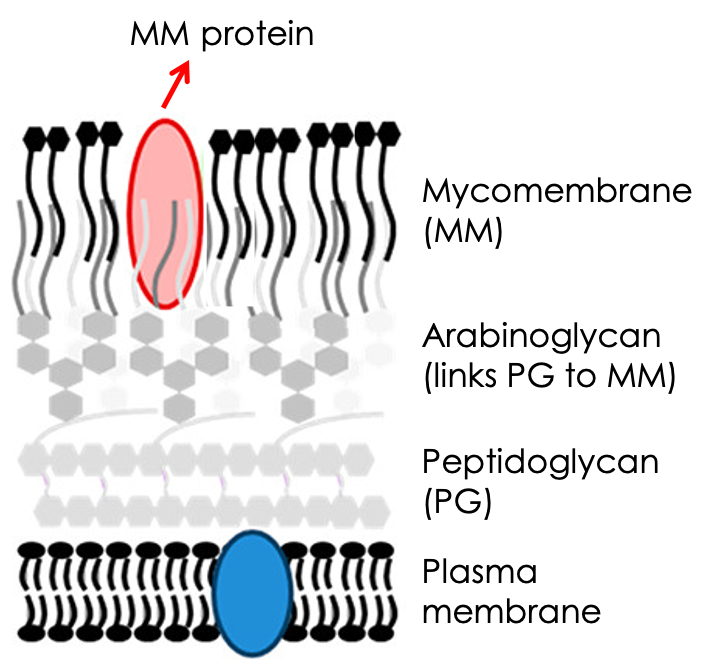
Significant effort has been funneled to elucidating the composition and formation of the mycomembrane, as it could guide the development of therapeutics against these pathogens. Indeed, several tuberculosis drugs work precisely by compromising the bacterium’s mycomembrane biosynthetic pathways. Great strides have been made in this regard in the past decades, but we still lack a full understanding of all the proteins involved in constructing the mycomembrane. Facing this challenge, the team of Benjamin Swarts at Central Michigan University proposes a chemical proteomics approach to find such elusive proteins, particularly those that interact directly with the mycolic acid ester subunits that make up the mycomembrane (Figure 2). Their approach relies on the synthesis of a photoactive version of a mycomembrane glycolipids known as TDM and AGM. The key to these probes is the presence of i) a diazirine moiety that generates a highly reactive species upon UV irradiation and ii) an alkyne group in the lipid’s tail that provides chemical reactivity to “click” additional molecules onto it.
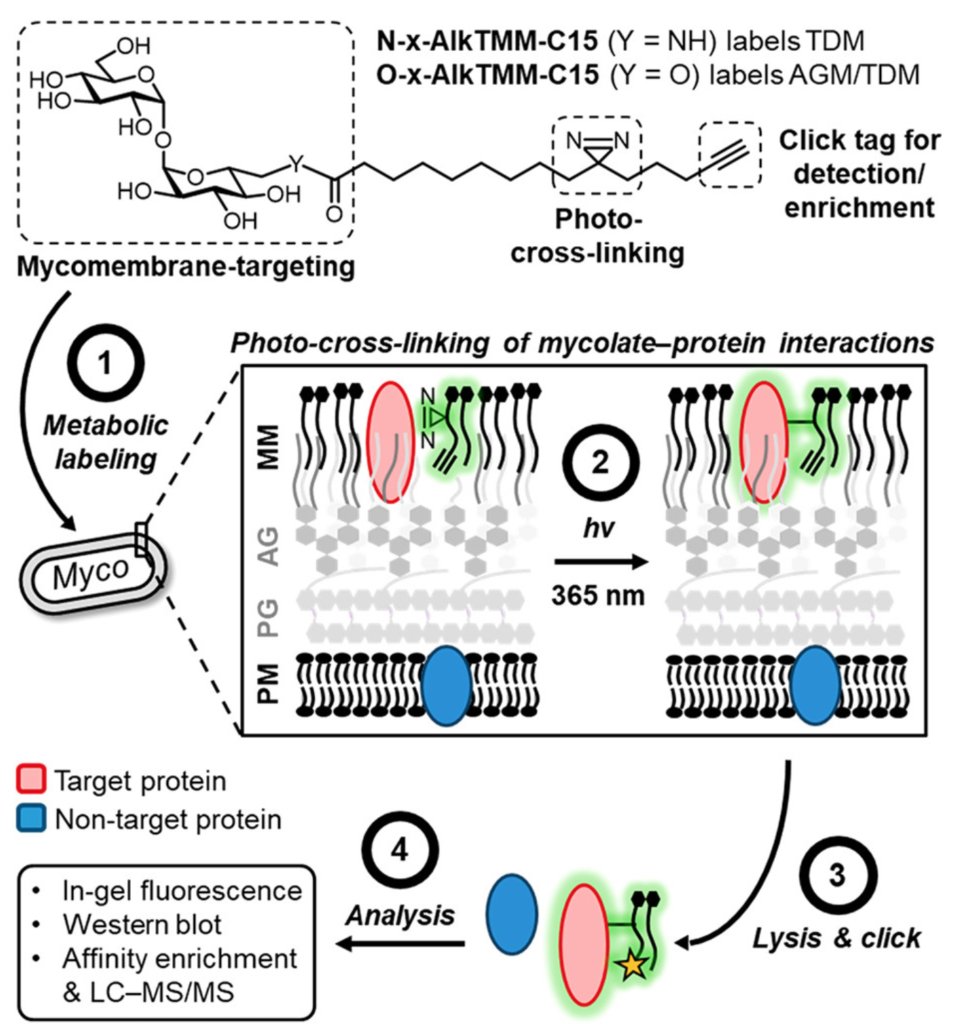
Even with these additions, these molecules are similar enough to their original counterparts. Thus, when fed to the non-pathogenic model Mycobacterium
Through chemical synthesis, the authors first got their hands on the diazirine-containing probes that would eventually label TDM or AGM. Before jumping into in vivo studies, they first confirmed the molecules perform well in vitro. They incubated the probes with a model protein called BSA, performed the photo-crosslinking, “clicked” a fluorescent dye to the alkyne tail, and ran the mixture on a gel to visualize, via in-gel fluorescence, if BSA got labeled.
The group then tested the incorporation of their mycolate probes in the mycomembrane of Msmeg, feeding either compound to the bacteria, “clicking” a dye to the alkyne, and assessing the incorporation levels by fluorescence microscopy. They observed good levels of dose- and time-dependent fluorescence in the membranes of Msmeg and another mycomembrane-containing bacterium. No signal was observed when feeding to mycomembrane-deficient specimens like E. coli or B. subtilis, highlighting the probe’s specificity (Fig. 3).
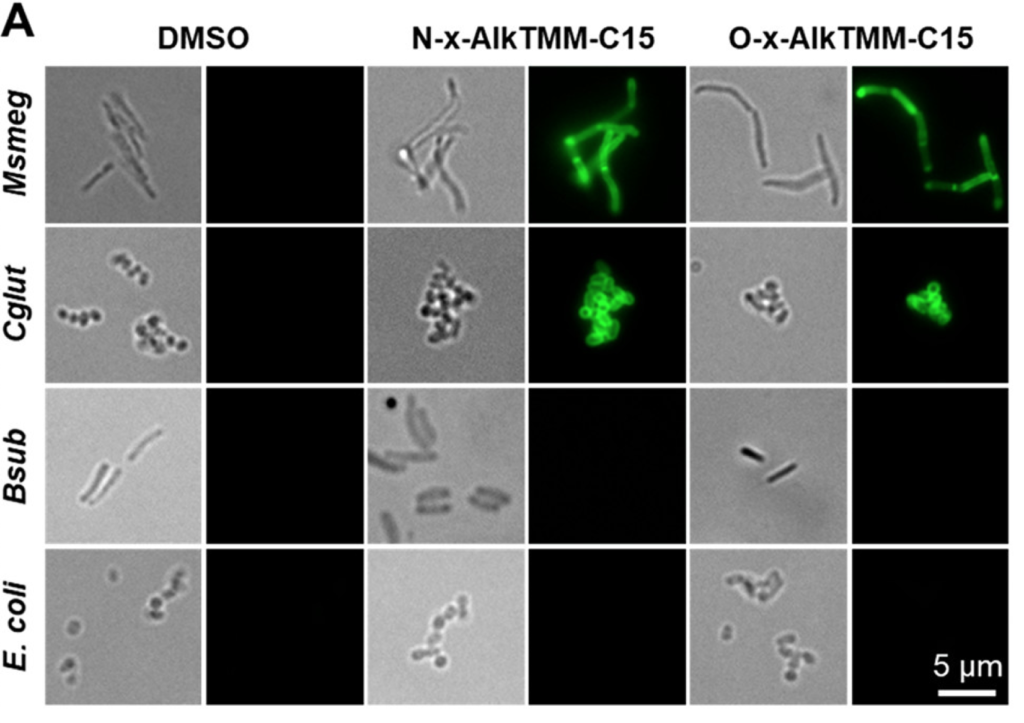
from Swarts, et al. JACS 2020, 142, 7725.
Finally, the team proceeded with the proteomic experiments, where bacteria with the modified mycomembranes were UV-irradiated and lysed. This time, the alkyne group was used to anchor any probe-linked adducts to a bead, essentially enriching for any proteins that were directly interacting with that glycolipid. The MS proteomics results revealed a panel of almost 100 proteins highly enriched by photo-crosslinking, including proteins implicated in mycomembrane biosynthesis/remodeling and other promising interactors uncharacterized in this regard, including the porin protein MspA.
These results are quite illuminating and hint at the extraordinary complexity of mycomembrane assembly and dynamics. Given the fundamental nature of this report, no biological follow-up was performed with any of the possible “hits,” so an ocean of questions remains unanswered. Still, this platform developed by Swarts and coworkers – the first ever to capture mycomembrane lipid-protein interactions in vivo – is a remarkable tool to map this complex interactome and can be adapted to other mycolipids from other organisms, thus adding to our understanding of mycobacterial physiology and pathogenicity. Who knows, maybe future mappings will reveal suitable targets to fight the next mycobacterial infections.