Title: “Chemical Control of Quorum Sensing in E. coli: Identification of Small Molecule Modulators of SdiA and Mechanistic Characterization of a Covalent Inhibitor”
Authors: Matthew J. Styles, Stephen A. Early, Trisha Tucholski, Korbin H. J. West, Ying Ge, Helen E. Blackwell
Journal: ACS Infectious Disease
Year: 2020
https://pubs.acs.org/doi/abs/10.1021/acsinfecdis.0c00654
Featured image and figures used with permission. Further permissions related to this material must be directed to the ACS.
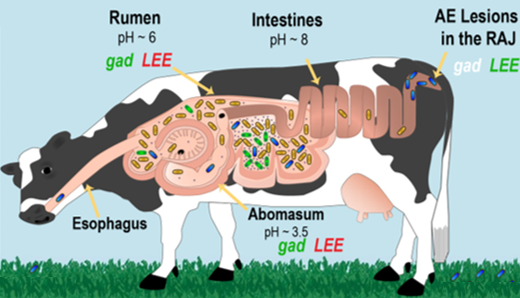
If you have ever cooked or eaten ground beef, you may have been cautious about making sure your burger or meatloaf is cooked completely through. This thorough cooking is done to kill harmful bacteria—particularly Escherichia coli (E. coli)—that can live in the intestines of cattle and contaminate the meat during processing [1]. Some E. coli can produce toxins that make humans incredibly sick, with severe diarrhea and intestinal bleeding [1]. While many different strains of E. coli exist, enterohemorrhagic E. coli (EHEC) is the strain that most commonly causes severe illness in humans due to the Shinga toxin it produces [1]. Although 75% of EHEC infections in humans originate from infected cattle, there are still a lot of unknowns about how EHEC survives and thrives in the digestive tract of cows. In their recent work, Styles and coworkers sought to find a way to stop these bacteria from getting to and damaging the intestines by preventing the bacteria from surviving the acidic environment of a cow’s stomachs.
Because there is strength in numbers, EHEC and many other pathogenic bacteria communicate with each other through a method called quorum sensing (QS) where individual bacteria produce and secrete signaling molecules into their environment. When many bacteria are crowded in one place, these signaling molecules accumulate enough to be detected by bacterial receptors. These receptors often act as switches to turn ON the expression of genes encoding virulence factors, which are molecules that enable the bacteria to both colonize the host and cause disease. In EHEC bacteria, the receptor being studied by Styles and coworkers is called SdiA (Fig. 1A), and it can detect a whole family of molecules commonly used by Gram-negative bacteria for quorum sensing, called N-Acyl L-homoserine lactones (AHLs, Fig. 1B).
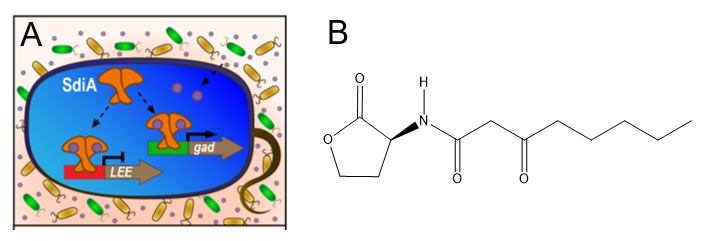
In the cell, two SdiA receptors bound to AHL molecules dimerize (come together to form a pair) and are then able to turn ON the expression of the gad genes that allow the bacteria to survive in acidic environments like cow stomachs. Interestingly, the SdiA/AHL dimer also turns OFF the LEE genes that allow the EHEC to create lesions in the digestive system. This makes sense, however, because unlike the stomachs which are acidic at pH 6 and pH 3.5, the cow intestines are alkaline at pH 8 (Fig. 2A). The “lactone” group of N-acyl L-homoserine lactones is actually hydrolyzed (broken apart by a water molecule) in alkaline environments, so in-tact AHL molecules are not available to bind SdiA receptors and turn LEE genes OFF. Therefore, the LEE genes produce virulence factors that create lesions in the intestine, causing disease symptoms.
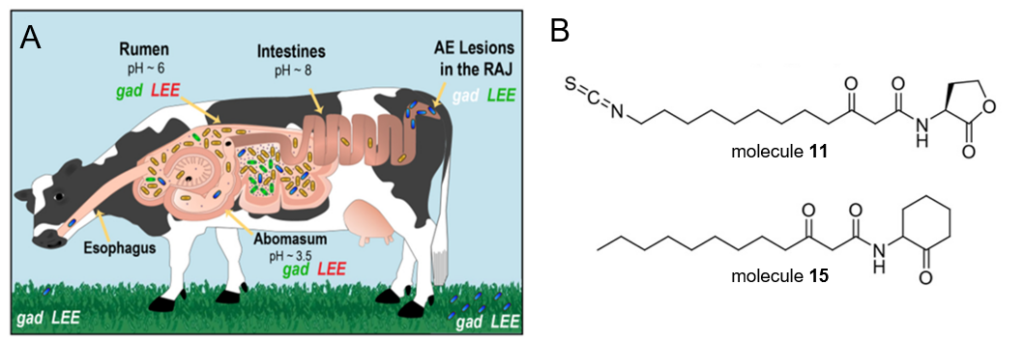
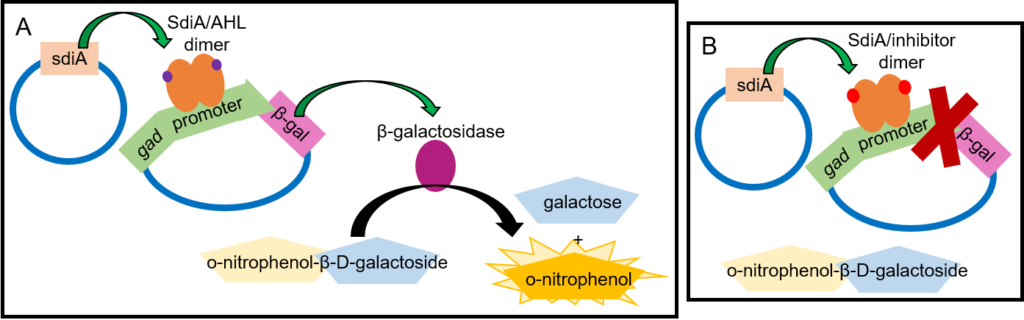
Despite the importance of SdiA in EHEC survival and virulence, scientists had never identified any SdiA inhibitors. Styles and coworkers sought to solve this problem by testing the ability of analogues (similar molecules, Fig. 2B) of AHLs to bind SdiA receptors but prevent the receptors from turning gene expression ON and OFF. In their initial experiment, the scientists used a standard test called a β-galactosidase assay (Fig. 3) which requires two plasmids (circular pieces of DNA that bacteria can use), a synthetic compound called o-nitrophenol-β-D-galactoside, and the analogue to be tested, which are all combined in harmless lab bacteria [2]. One plasmid has the gene that encodes SdiA so the bacteria can make the receptor [2]. The other plasmid contains the DNA sequence that the SdiA/AHL dimer usually interacts with to turn the gad genes ON, but the gad genes themselves have been replaced with the gene for the β-galactosidase enzyme [2]. β-galactosidase then breaks the synthetic o-nitrophenol-β-D-galactoside molecule down into galactose and a yellow molecule called o-nitrophenol [2]. In this way, a normal AHL signaling molecule would form the SdiA/AHL dimer and turn ON the expression of β-galactosidase, and a yellow color would be visible. An inhibitor, however, would result in no yellow color. To be sure that their potential analogues were actually inhibiting SdiA rather than just not interacting with SdiA at all, the scientists performed the assay with a normal AHL signaling molecule combined with an analogue to see if the analogue would prevent the real signal from working to produce that yellow color. Happily, this assay identified two inhibitors, named molecule 11 and molecule 15 (Fig. 2B).
Finally, the scientists tested whether molecules 11 and 15 could prevent EHEC survival in acidic conditions. For this experiment, the scientists grew the EHEC in a liquid broth at neutral pH, then split the EHEC into new broth that was either neutral (pH 7.0) or acidic (pH 2.5) for 1.5 hours. They then calculated acidic environment survival as a percentage of EHEC living in the acidic broth relative to the EHEC living in the neutral pH broth. They repeated this procedure with an EHEC mutant where the sdiA gene was deleted (∆sdiA). Lastly, they repeated this procedure again with each inhibitor. Happily again, they found that both 11 and 15 decreased the acidic environment survival of the wild type (non-mutant) EHEC down to almost the same levels as the ∆sdiA mutant, showing that each inhibitor successfully prevented normal function of the SdiA receptor.
More experiments would still be required to determine whether molecules 11 and 15 could one day be used as medication to treat cattle or especially humans with EHEC infections, since potential drugs often have hazardous side effects. Additionally, the role of SdiA in turning the LEE genes OFF might complicate the ability to use an SdiA inhibitor to prevent infection. However, this discovery of the first two known EHEC SdiA inhibitors is an important first step toward one day designing a molecule to prevent the virulence factors that make EHEC such a dangerous pathogen.
[1] https://www.hopkinsmedicine.org/health/conditions-and-diseases/enterohemorrhagic-escherichia-coli
[2] https://openwetware.org/wiki/Beta-Galactosidase_Assay_(A_better_Miller)