Title: PEARL-Seq: A Photoaffinity Platform for The Analysis of Small Molecule-RNA Interactions
Authors: Jennifer C. Petter, et al.
Journal: ACS Chemical Biology
Year: 2020
https://pubs.acs.org/doi/10.1021/acschembio.0c00357
When one thinks of developing therapeutics, it is often with the proteome in mind. After all, the ability of proteins to regulate a myriad of cellular processes is finely tuned by the tertiary structures they adopt, making them the obvious targets for small molecule “drugging.” More recently, however, RNA is emerging as a new focus for novel therapeutics. Much like proteins, RNAs can oversee important cellular events, such as transcription and translation. Moreover, scientists have shown that these regulatory functions often hinge on how these RNAs fold, injecting interest into the area of small molecule-RNA targeting.
Some research already exists in examining the interactions of known small molecule drugs and key RNA molecules. Yet, deploying a more expansive approach, particularly one where prior knowledge of possible RNA targets is not needed, will require high throughput methods to map, confirm, and characterize novel small molecule-RNA binding events. A group led by Jennifer Petter at Arrakis Therapeutics has risen to the challenge by developing PEARL-Seq, a platform to pinpoint small molecule binding sites on RNAs. This technology capitalizes on photoaffinity labeling to stabilize and thus confidently identify sites of ligand-RNA interactions.
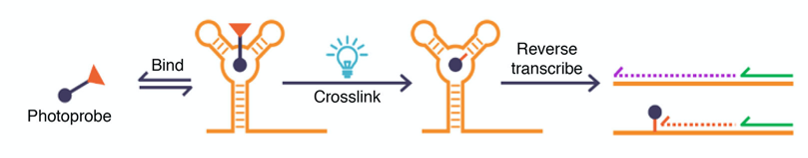
In short, a known small molecule candidate is functionalized with a photo-reactive group that exposes a highly reactive residue when activated with light. The compound is allowed to reversibly bind to its RNA target(s), but upon UV light irradiation, the reactive moiety is unmasked, and the molecule forms a covalent bond with the nearby RNA and makes the binding irreversible. The RNA is then reverse-transcribed (RT) in vitro to make a complementary DNA (cDNA) copy, but sites where crosslinking occurred induce a stop in the “copying” process, resulting in a truncated cDNA. Finally, sequencing of this cDNA reveals the truncation sites and yields the exact spots where small molecule targeting occurred (Figure 1).
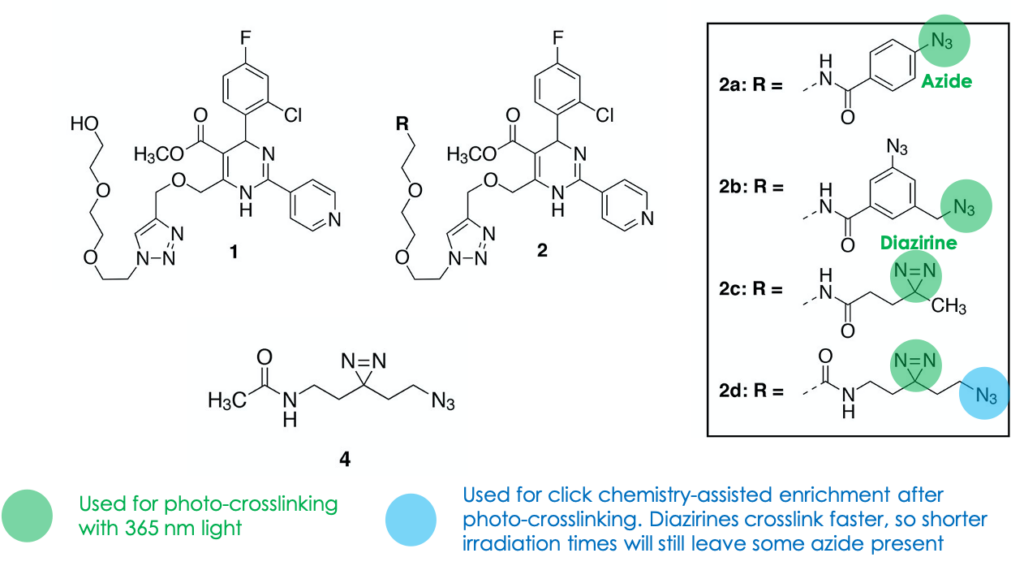
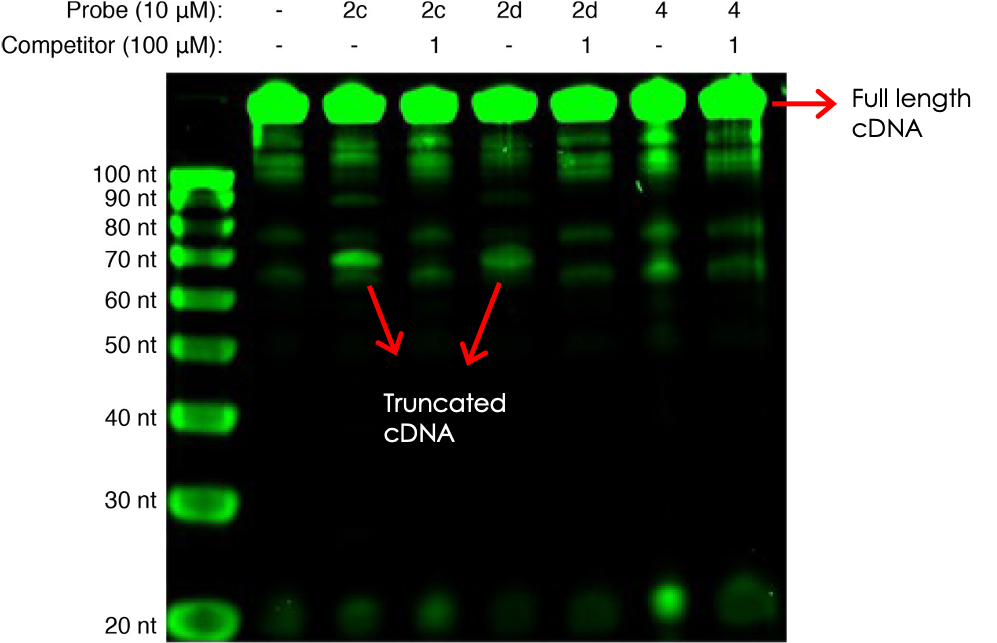
The authors first established proof-of-concept by studying a model system: the binding of a known RNA molecule (named Aptamer 21) to small molecule 1. They synthesized photo-reactive variations of 1 that contained either azide or diazirine groups (molecules 2a–2d, Figure 2), attempted photo-crosslinking, and observed covalent adduct formation by mass spectrometry. Next, they confirmed that the crosslinking generated stops upon RT using an assay designed to generate a fluorescent cDNA copy that could be detected on a gel. Incubation with the photo-reactive candidates indeed resulted in site-specific truncations, as seen by shorter-running fluorescent bands on the gel (Figure 3). Competition for binding with the original ligand 1 restored the higher-running, full-length cDNA, confirming that crosslinking was dependent on ligand binding. Finally, sequencing of the RNA photo-adducts revealed a relatively limited number of truncation sites, supporting the interaction’s site-specific nature.
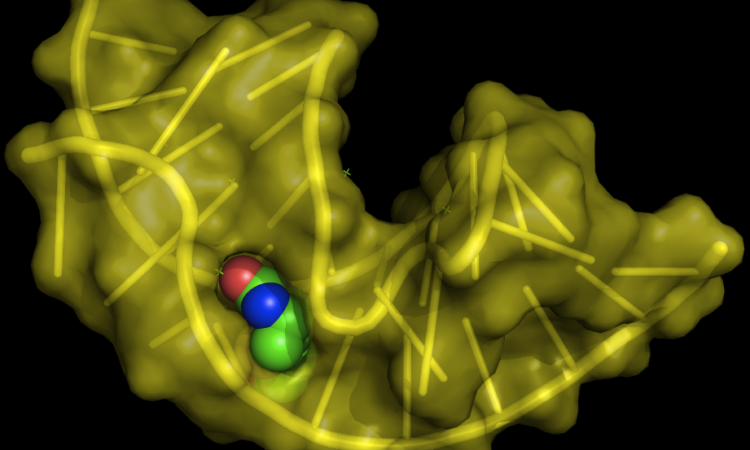
To test the platform’s selectivity under more demanding conditions, the group used probe 2d (Figure 2) to target Aptamer 21 spiked into a more complex mixture of cellular mRNA. The probe’s diazirine moiety was used for target crosslinking as usual, and the azide group functioned as a handle to “fish-out” the crosslinked adducts by click chemistry-aided enrichment (the azide can react orthogonally with an alkyne-containing affinity “bait”, such as a biotin-alkyne). Not only did this result in enrichment of Aptamer 21, highlighting the approach’s selectivity, but it also yielded crosslinking sites identical to those obtained on a simpler in vitro setup, confirming the technology operates well on more complex settings. To conclude, the group went on to interrogate the probes’ interactions with aptamer 21 using 2D-NMR experiments and molecular modeling and noted that sites of docking and contact are consistent with the truncation sports revealed by PEARL-Seq. I will leave the more intricate details out but encourage the curious reader to refer to the paper to find more.
While the authors did not advance the method beyond their model Aptamer 21 system, their achievement is commendable. In this more advanced iteration, not only is PEARL-Seq able to pin down and enrich form a complex mixture the RNA binding partners of a given small molecule probe, but also enrich and inform on the molecule’s precise binding site. One can envision applying this approach to elucidate the mechanism of action of a specific RNA-binding ligand or find RNA off-targets of known or potential dugs. Coupled with structural probing RNA and structure-activity relationship (SAR) assays that are routine in the therapeutic world, PEARL-Seq could even advance the rational design of new RNA-binding molecules.