Article Title: Plasmalogen Biosynthesis by Anaerobic Bacteria: Identification of a Two-Gene Operon Responsible for Plasmalogen Production in Clostridium perfringens
Authors: Jackson, D. R.; Cassilly, C. D.; Plichta, D. R.; Vlamakis, H.; Liu, H.; Melville, S. B.; Xavier, R. J.; Clardy, J.
Journal: ACS Chem. Biol.
Year: 2020
DOI: 10.1021/acschembio.0c00673
There is growing evidence that the human microbiota, which consists of trillions of microscopic bacteria, archaea, viruses, eukaryotes, plays an integral role in human health and disease. It is often dubbed to be an “essential organ” and collectively has 150 times more genes than the human genome. As a result, there is great curiosity to understand what enzymes these microbial genes encode and what compounds these enzymes produce. In this paper, Jackson et al. focus on how the microbiota produces a set of compounds that are part of the plasmalogen family of lipids.
Plasmalogens are found across bacteria and animals, including human brain, heart, and immune cell membranes. They play diverse roles in signaling, structure, and protection against oxidative damage; the lack of plasmalogen biosynthesis can result in terminal diseases such as rhizomelic chondrodysplasia punctata. The characteristic chemical feature of plasmalogens is a long-chain vinyl ether attached to a glycerol molecule (Figure 1). Although an aerobic, O2-dependent pathway for producing plasmalogens is known, this is not relevant in the anaerobic human gut microbiota environment where O2 is not present. To fill this gap in knowledge, the researchers studied the plasmalogen-producing anaerobe (microbe that does not require O2) Clostridium perfringens to uncover the anaerobic pathway.

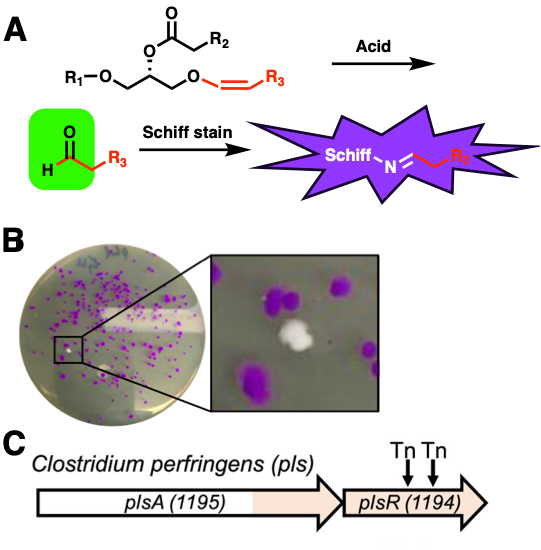
The researchers took advantage of two things: a colorimetric assay to screen for plasmalogen production and a previously constructed C. perfringens transposon library. Transposon libraries are collections of multiple cell mutants with different individual genes rendered non-functional. These are often used to screen and identify the role of individual genes. In addition, researchers previously noted that when plasmalogen-producing cells are treated with acid, a reactive aldehyde species forms that can react with Schiff stain and turn cells a characteristic purple color (Figure 2A). Using these two concepts, the researchers screened the library for cell mutants that did not turn purple on an agar plate. Cells that remained white could not produce plasmalogens and likely have mutations in genes important for plasmalogen production (Figure 2B). By sequencing these white cells, two C. perfringens genes, plsA and plsR, were identified (Figure 2C).
To confirm that these two genes alone were enough to produce plasmalogens, plsA and plsR were introduced into Escherichia coli, a microbe that does not naturally make plasmalogens. Satisfyingly, plasmalogens were detected only when the genes were present and when the microbe was grown in the absence of O2. Further investigation of the gene sequences showed that they are related to characterized enzymes of the BCR/HAD family, a subset of enzymes that use one-electron chemistry to catalyze very challenging reductive reactions that cannot be accomplished by common biological reductants (e.g. NAD(P)H, flavins, hydroquinones). This allowed the researchers to propose a mechanism by which plasmalogens are formed under anaerobic conditions, although additional follow studies will be needed.
Other than expanding the known repertoire of BCR/HAD enzymes, Jackson et al. investigated how widely-distributed these genes are in the human microbiome. In the end, they found 1910 microbiome-associated species. These included facultative anaerobes (microbes that can grow both with and without O2), which were previously believed to not be able to produce plasmalogens. One such example is Enterococcus faecalis, a common gut microbe. The researchers grew E. faecalis on agar plates in the presence and absence of O2 and screened for plasmalogen production using Schiff staining as described above. Interestingly, the anaerobically grown microbes were completely purple while aerobically grown microbes had a white exterior and purple interior. O2 is limited in the center, which can explain why there is a color, and thus plasmalogen production, difference.
Through this work, an O2-independent method of producing the important class of molecules, plasmalogens, was identified. The two relevant genes are part of an understudied enzyme family that catalyze challenging reductions and opens the door to additional biochemical characterization. In addition, these genes are widely present in the human microbiota, including microbes previously thought to not be able to produce plasmalogens. It will be interesting to see how disentangling host- and microbiota-derived plasmalogens increases our understanding of the complex interactions of human microbiota.