Article Title: Co-occurrence of enzyme domains guides the discovery of an oxazolone synthetase
Authors: de Rond, T.; Asay, J. E.; Moore, B. S. et al.
Journal: Nat. Chem. Biol.
Year: 2021
DOI: 10.1038/s41589-021-00808-4
With the advent of next generation DNA sequencing technologies, large amounts of genetic information can be obtained from diverse environments including the ocean, deep thermal vents, and the human gut microbiome. The vast majority of these encoded genes have never been sequenced before, meaning the functions of these enzymes are unknown. These uncharacterized enzymes catalyze novel chemical transformations that could be used to understand basic biology or used in biocatalysis cascades to mass produce chemicals. Most approaches to annotate genes rely on recognizing sequence homology to known enzymes. For instance, if an unknown protein is 80% identical to a characterized enzyme, it likely catalyzes the same reaction. These methods tend to rediscover known enzymes rather than expand the chemical toolbox. To address these limitations, de Rond et al. developed a co-occurring enzyme domains (CO-ED) workflow to discover enzymes.
Enzymes with specialized roles often possess multiple catalytic domains (a stretch of the enzyme that binds substrate and catalyzes a reaction). Unlike most enzymes that contain one domain and catalyze a single chemical transformation, these multi-domain enzymes can catalyze two, and sometimes dozens of, sequential reactions. The authors devised a workflow to identify enzymes with co-occurring enzyme domains (CO-ED) that had never been seen together before. Briefly, this involves generating a list of enzymes of interest, identifying what domains are present, and graphically representing the network (Figure 1). By looking at the network, uncharacterized CO-EDs can quickly be visualized and prioritized for further experimental study.
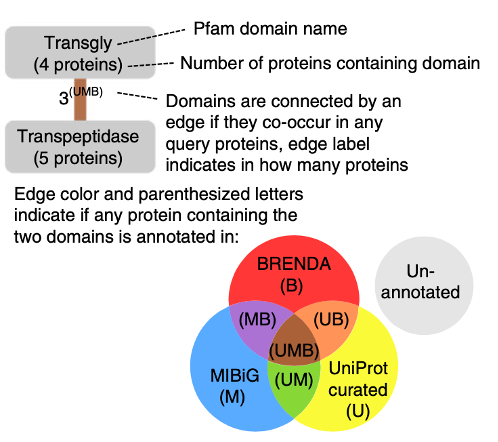
To validate this workflow, the authors queried the genes from the commonly used lab-strain Escherichia coli K12. There were 19 co-occurring domains, all connected with a colored edge, implying the functions of these enzymes were known. On the other hand, when this same method was used with a less well-studied γ-proteobacterium Pseudoalteromonas rubra DSM 6842, there were 12 co-occurring domain-pairs that had never been seen before.
The authors decided to focus on the domain pair “ThiF”–“nitroreductase,” as both domains individually catalyze a large variety of transformations. Importantly, all known substrates of enzymes with ThiF-domains are polypeptides or tRNAs. This gene was named OxzB and could be found in a variety of other Proteobacteria alongside the N-acyltransferase enzyme OxzA. When these two genes were heterologously expressed in E. coli, the cells turned visibly yellow. Using a combination of mass spectrometry, liquid chromatography, and NMR spectroscopy, the products of OxzAB were identified to be a series of oxazolones (Figure 2).
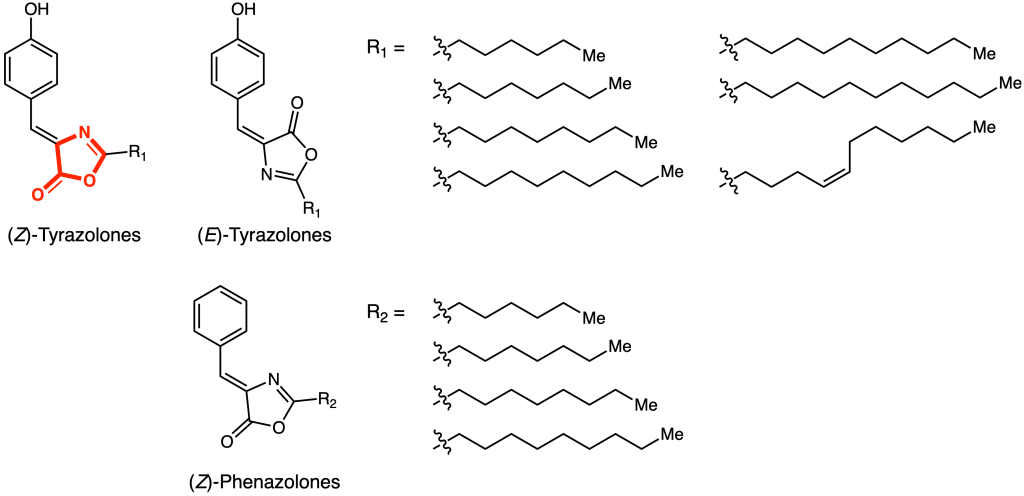
Moreover, the enzymes could be individually expressed and purified, and tested for activity in vitro. Indeed, incubation of L-tyrosine and decanoyl-CoA with OxzA led to formation of N-decanoyl-L-tyrosine. Further reaction with OxzB led to formation of the (E)-tyrazolone, which isomerized over time to form (Z)-tyrazolone (Figure 3). As a final sanity check, the authors were able to elicit and detect production of these compounds in the native Proteobacteria species.

Through this CO-ED approach, de Rond et al. were able to show as a proof-of-concept that there are many novel multi-domain enzymes waiting to be characterized. These likely catalyze diverse reactions, such as OxzB discussed here. Although this is not the first oxazolone-producing enzyme discovered, it is the first to act on small molecules rather than polypeptides, making it more amenable for latter biocatalysis efforts. It will be exciting to see how other labs around the world use this new approach, especially since not all enzymatic products have such explicit phenotypes (e.g., turn cells yellow). Nevertheless, this is a nice method for prioritizing study of more diverse enzymes.