Title: High-Throughput miRFluR Platform Identifies miRNA Regulating B3GLCT That Predict Peters’ Plus Syndrome Phenotype, Supporting the miRNA Proxy Hypothesis
Authors: Chu T. Thu, Jonathan Y. Chung, Deepika Dhawan, Christopher A. Vaiana, and Lara K. Mahal
Journal: ACS Chemical Biology
Year: 2021
https://pubs.acs.org/doi/10.1021/acschembio.1c00247
Featured image and figures reproduced with permission. Further permissions related to this material should be directed to ACS.
If you have ever learned about cell biology or molecular biology in school, you probably learned about the Central Dogma which states that genes encoded in DNA are transcribed into messenger RNAs (mRNAs) which are then translated into proteins that carry out jobs within the cell, thus turning genetic information into a functioning, living organism. While the Central Dogma is a good starting point, the life of an actual cell turns out to be a lot more complicated, with multiple different kinds of regulators that either increase or decrease transcription or translation. One kind of regulator is the microRNAs (abbreviated miRNAs or miRs) which are short RNA strands that are not translated, but instead bind non-covalently to specific mRNAs to control the mRNA expression. miRNAs can work in multiple different ways including: decreasing translation, increasing translation, and recruiting enzymes that degrade mRNA, as well as other mechanisms [1]. Although it is currently possible to purchase a library of all known human miRNAs (Sigma sells a library containing 2,754 miRNA mimics for over $9,000) [2], it is still mostly unknown how each specific miRNA works and even which genes each miRNA targets. Recently, scientists Chu and coworkers decided to find out which human miRNAs regulate the gene expression for beta-3-glucosyltransferase (B3GLCT) because mutations in this gene are known to cause Peter’s Plus Syndrome: a congenital (present at birth) disorder with a range of possible symptoms including cleft palate, intellectual disability, and abnormalities in the brain, facial structure, height, and limbs.
B3GLCT is a glycosylation (“sugar adding”) enzyme that attaches sugars to thrombospondin type-1 repeat (TSR) proteins. TSRs play an important role in embryo development [3], so it makes sense that developmental abnormalities would occur when TSRs lack their necessary sugar attachments. To research the effect of miRNAs on B3GLCT expression, Chu and coworkers began by purchasing the human miRNA library from Sigma. They then used computational prediction software to predict which miRNAs might target the B3GLCT mRNA. One such computer program called Targetscan 7.2 predicted that 480 out of the 2,754 miRNAs would target B3GLCT. However, Chu and coworkers knew that miRNA-target interactions predicted by computers tended to have high rates of both false positives and false negatives. More experimental data examining miRNAs and their targets is still needed to improve the predictive power of these programs.
One reason for the lack of miRNA data is the lack of a good assay for screening many miRNAs against one target gene in a living cell. For this reason, Chu and coworkers developed a simple miRNA-target assay that they named miRFluR (miR Fluorescent Ratiometric assay, Fig. 1). In this assay, Chu and coworkers constructed a plasmid (a circular piece of DNA) containing the gene for a blue fluorescent (light-emitting) protein called Cerulean and the gene for a red fluorescent protein called mCherry. Directly after the Cerulean gene, Chu and coworkers added the region of the B3GLCT gene most likely to bind miRNAs (the region is called the 3´-untranslated region, or 3´-UTR). They inserted this plasmid along with each of the 2,754 miRNAs into a line of human cells called HEK-293T, such that each sample would have the same plasmid but a different miRNA. They also included one positive control, miR-200b-3p which is already known to target the B3GLCT 3´-UTR, and two negative controls, miR-200a-3p and a nontargeting control (NTC) which are known to not target B3GLCT. They tested each miRNA in triplicate (three times).
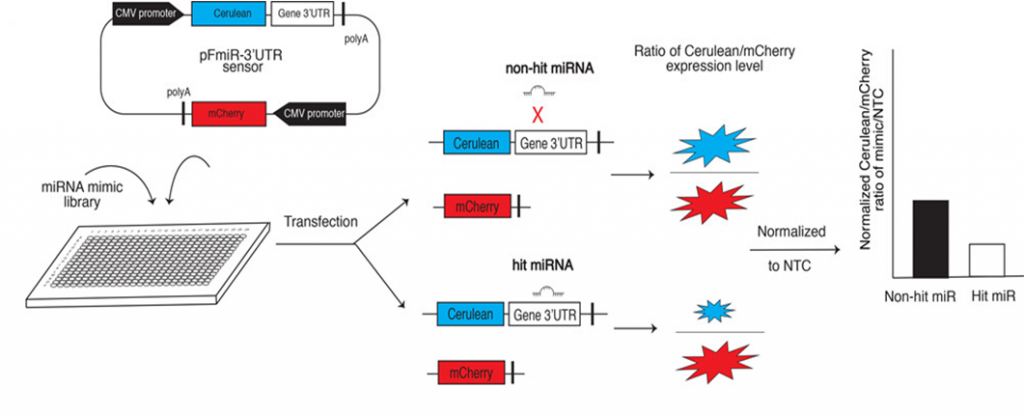
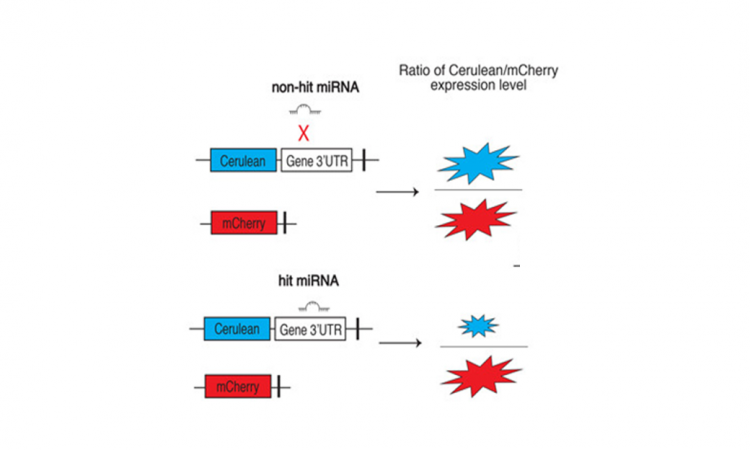
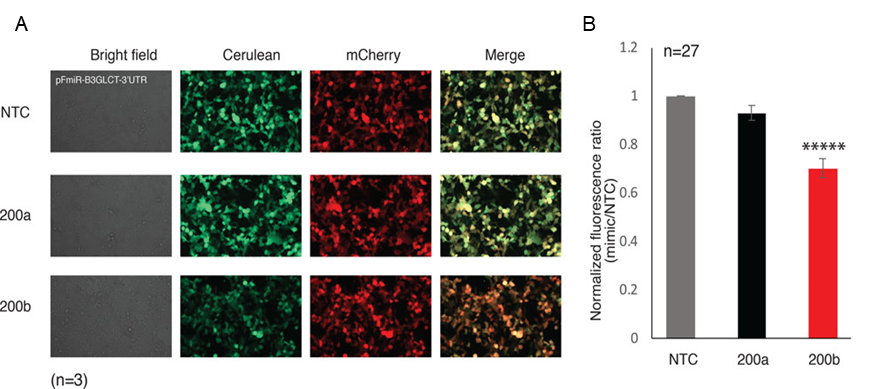
In the assay, both Cerulean and mCherry proteins are produced by the HEK-293T cells, causing both blue and red light to come from the cells (Fig. 2). If a miRNA targets the B3GLCT 3´-UTR to decrease expression, then the scientists expected to see less blue light, so the ratio of blue to red light should decrease compared to the NTC control (Fig. 1). Chu and coworkers first tested this method with only their positive and negative controls, and they did see a decrease in the blue to red ratio for the positive control miR-200b-3p, but not the negative control miR-200a-3p (Fig. 2).
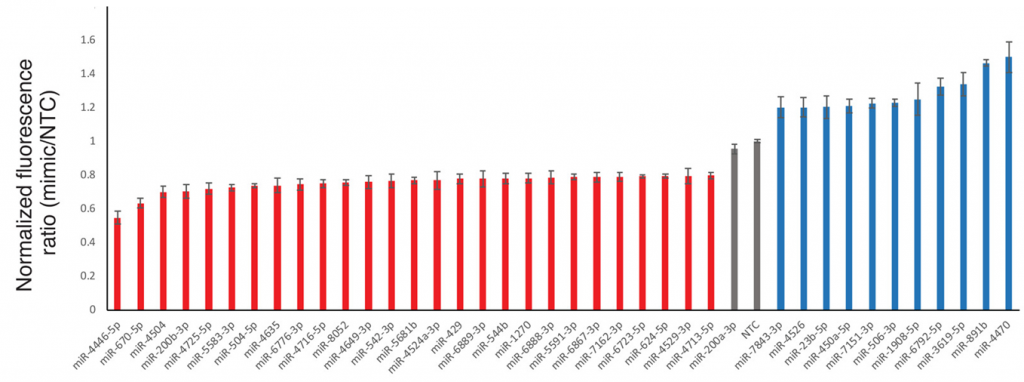
Once their method was developed and tested, the scientists were ready to test all the miRNAs in the library. To be consistent with previous miRNA research, Chu and coworkers defined downregulation (a decrease in expression) as a 20% decrease in the blue to red ratio compared to the NTC control, and upregulation as a 20% increase. Overall, they found 27 miRNAs to be downregulators of B3GLCT expression and 11 miRNAs to be upregulators (Fig. 3). In comparison, Targetscreen predicted only about 2/3 (17/27) of the downregulators, and none of the upregulators. Out of the 480 Targetscan predicted downregulators, only 3.5% actually downregulated BL3GLCT expression in the experiment. This shows how important it is for scientists to collect more data in the lab to better understand which miRNAs regulate the expression of individual genes.
Overall, Chu and coworkers have developed a useful assay for testing many miRNAs against one target gene, while simultaneously shedding light on the accuracy of miRNA-target prediction software. They have also identified 37 new miRNAs that may be useful in further investigations of B3GLCT expression and Peter’s Plus Syndrome. Although some individuals Peter’s Plus Syndrome have completely nonfunctional versions of B3GLCT, others have partially functioning versions [5]. For this reason, understanding how B3GLCT expression is regulated could potentially lead to the development of new therapies for some individuals.
[1] https://portlandpress.com/biochemsoctrans/article/36/6/1224/66536/How-do-microRNAs-regulate-gene-expression
[2] https://www.sigmaaldrich.com/US/en/product/sigma/mi00300
[3] https://anatomypubs.onlinelibrary.wiley.com/doi/10.1002/(SICI)1097-0177(200006)218:2%3C280::AID-DVDY4%3E3.0.CO;2-0
[4] https://www.nature.com/articles/nrd.2017.116
[5] https://rarediseases.info.nih.gov/diseases/8422/peters-plus-syndrome