Title: Methylation-Sensitive Loop-Mediated Isothermal Amplification (LAMP): Nucleic Acid Methylation Detection through LAMP with Mobile Fluorescence Readout
Authors: Jacob Amos Hambalek, Janay Elise Kong, Calvin Brown, Hector Enrique Munoz, Thomas Horn, Michael Bogumil, Eleni Quick, Aydogan Ozcan, and Dino Di Carlo
Journal: ACS Sensors
Year: 2021
Figures adapted with permission from Hambalek et al. ACS Sensors Article ASAP. Copyright 2021 American Chemical Society.
https://doi.org/10.1021/acssensors.1c00902
Although the genes coded into our DNA can have a strong effect on our traits—blue eyes or brown? does cilantro taste delicious or like soap?—there is more to the story than just variation or mutation in the genes themselves. Our DNA’s genetic code can be thought of like a cookbook full of recipes. However, our cells have to decide which recipes to make, how much, and how often. This is how cells in our eyes and cells in our skin can perform different functions, even though all the cells in our body have the same DNA. The process of making the “recipe” of a specific gene is known as gene expression, and usually involves transcription of a gene into messenger RNA (mRNA) followed by translation of the mRNA by the ribosome to make a protein that does jobs for the cell. One way the cell can decrease transcription of a specific gene is through a process called DNA methylation where a methyl group (a carbon atom bound to three hydrogen atoms) is chemically attached to the DNA at a specific site. DNA methylation usually occurs in the promoter sequence next to a gene, since this promoter controls whether the gene is transcribed or not.
DNA is a double-stranded helix made up of four nucleotides: cytosine (C), guanine (G), adenine (A), and thymine (T), and methylation frequently occurs on a cytosine nucleotide followed by a guanine nucleotide. This is called CpG methylation and is an important marker in some diseases, especially in cardiovascular disease and some cancers. Detection of CpG methylation could potentially be used to diagnose disease, including to determine which kind of cancer a person has. Especially because some cancers are fairly resistant to chemotherapy, a quick and cost-effective CpG methylation detection method could help doctors diagnose and treat cancer and other diseases more effectively. For this reason, the scientists Hambalek and coworkers decided to develop a CpG methylation detection method that could hopefully be used in doctor’s offices in the future.
As their initial target DNA sequence, the scientists chose the BRCA1 promoter because it is found to be highly methylated in very aggressive types of breast cancer. First, the scientists isolated all the DNA from two different breast cancer cell lines: HCC38 which is known to have a lot of DNA methylation and MCF7 which has almost no methylation. Then, without any other sample preparation needed, they were able to put this isolated DNA through their two-step methylation detection protocol to see if they could differentiate between the two cell lines.
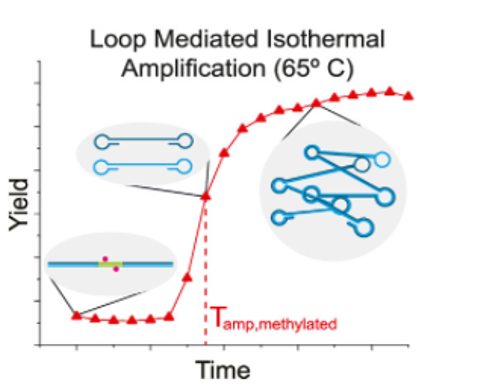
The first step (Fig. 1A) is to incubate the DNA with Methylation-Sensitive Restriction Endonucleases (MSREs) which are enzymes that cut the DNA helix like a pair of scissors, but only when methylation isn’t present. The scientists hypothesized that the methylation-lacking DNA from MCF7 cells would be cut up by the MSREs, while the HCC38 DNA would remain intact, protected from the enzyme by its methyl groups. The second step (Fig. 1B) is more complicated to understand but is still inexpensive and easy to perform, which would be important for use at a doctor’s office. This step is called Loop-mediated isothermal Amplification (LAMP). A video explaining this method is available from New England Biolabs: https://www.neb.com/tools-and-resources/video-library/loop-mediated-isothermal-amplification-lamp-tutorial?autoplay=1
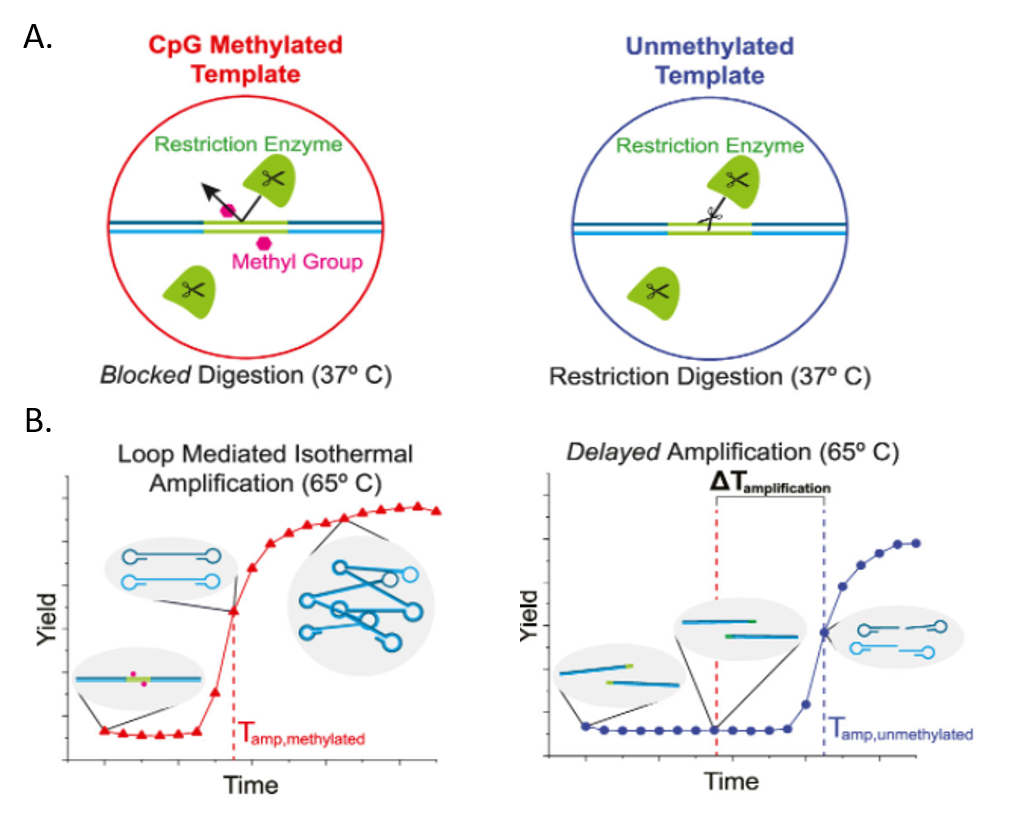
The LAMP method uses an enzyme called DNA polymerase which can make a copy of a DNA sequence. To copy the sequence, DNA polymerase reads one strand of the DNA double helix and takes nucleotides floating in the solution to build the complementary strand—where A is complementary to T, and G is complementary to C. Importantly, however, DNA can’t begin a complementary strand on its own. A small section of the complementary strand has to be present first, and this small piece of specific DNA is called a primer. For their experiment, Hambalek and coworkers designed primers that are complementary to either end of the BRCA1 promoter so that the polymerase would copy only their target sequence. LAMP requires special primers that will loop back on themselves to form a dumbbell shape (Fig. 1B). Because the primers become part of the complementary strand, the complementary strand will also have a dumbbell shape. Then, one dumbbell can act as the primer for another dumbbell, and DNA polymerase can make an exponential number of products. The DNA samples lacking methylation, however, have been cut up by the MSREs, so DNA polymerase can only make half dumbbells with these samples. These half dumbbells can still be copied, but it takes longer to get to the same level of amplification from half dumbbells compared to the full dumbbells.
To measure the amplification level in each sample, the scientists use a fluorescent dye called EvaGreen which binds only to double-stranded DNA which is present only in the amplified dumbbells. EvaGreen only emits light (fluoresces) when it’s bound to DNA, so measuring the amount of light emitted from each sample can tell the scientists how much double-stranded DNA is present. Excitingly, the scientists did see that the highly methylated HCC38 samples emitted fluorescence more quickly than the non-methylated MCF7 samples (Fig. 2).
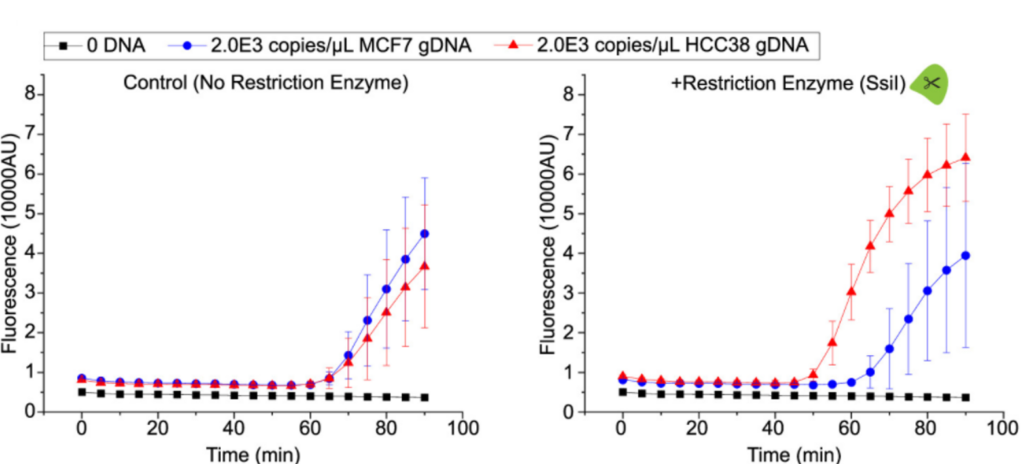
Fig. 2. LAMP fluorescence from the BRCA1 promoter. After incubation with MSRE Ssil, the methylated BRCA1 promoter from HCC38 cells (red) shows fluorescence sooner than the unmethylated promoter from MCF7 cells (blue). Without the MSRE step, both promoters give similar LAMP fluorescence, showing that MSRE is essential for this methylation detection method.
Overall, the scientists were able to differentiate between two cancer cell lines in just 80 minutes using inexpensive equipment. More work still needs to be done, however, before bringing this test to a doctor’s office. For example, more than two types of breast cancer exist, so additional tests of the BRCA1 promoter from more cancer cell lines are needed to determine just how powerful this technique could be. As a first step however, Hambalek and coworkers successfully created a method to measure disease-related methylation of DNA.
[1] https://www.tataa.com/wp-content/uploads/2012/10/EvaGreen-Manual.pdf