Article Title: New Role for Radical SAM Enzymes in the Biosynthesis of Thio(seleno)oxazole RiPP Natural Products
Authors: Lewis, J. K.; Jochimsen, A. S.; Lefave, S. J. et al.
Journal: Biochemistry
Year: 2022
DOI: /10.1021/acs.biochem.1c00469
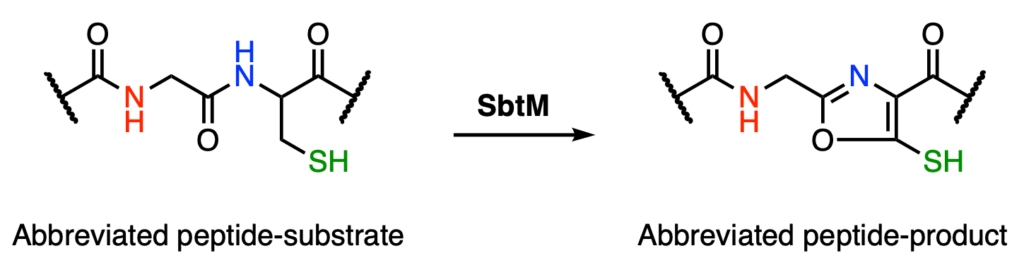
Beyond the production of primary metabolites required for life, microbes are capable of producing secondary metabolites known as natural products. These compounds are not essential for microbial survival, but often add an evolutionary benefit such as chemical targeting of prey, predators, and competing microbes. Ribosomally synthesized post-translationally modified peptides (RiPPs) are a class of natural products with diverse biological function. Typically, a peptide is synthesized by the ribosome and then modified by various enzymes to build chemical diversity. Radical S-adenosylmethionine (rSAM) enzymes are responsible for installing some of the most chemically important modifications, including complex cross links between specific amino acid residues. These include diverse C–S, C–C, C–O, and C–H bond-formation reactions. Here, Lewis et al. report the characterization of the rSAM enzyme SbtM, which catalyzes the cyclization of a cysteine or selenocysteine residue to form a thio(seleno)oxazole (Figure 1).
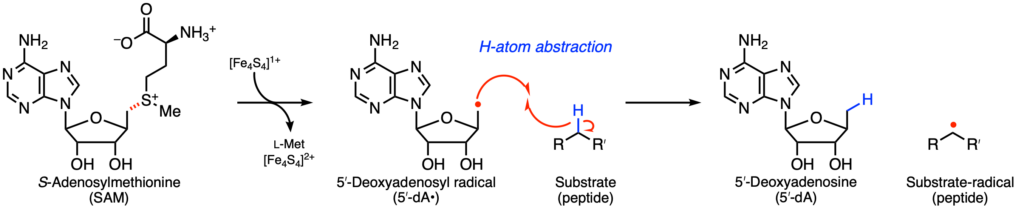
As their name implies, rSAM enzymes catalyze radical, or one-electron, transformations. These enzymes have a catalytic iron sulfur cluster [Fe4S4]1+ that reductively cleaves the cofactor SAM to form a 5′-deoxyadenosyl radical (5′-dA•). This transient, reactive species generates a substrate-based radical through hydrogen atom (H-atom) abstraction (Figure 2). Of all the uncharacterized rSAM enzymes, the authors were particularly interested in SbtM as it found in genomes near a peptide predicted to have a selenocysteine (SeCys) at position 35, so called the 21st amino acid. No SeCys-containing RiPP natural product has been characterized.
SbtM was expressed and purified, and the peptide was synthesized with either a Cys or SeCys residue at position 35. Both enzyme and either substrate were incubated together, and the reactions were analyzed by liquid chromatography-mass spectrometry (LC-MS). In both reactions, the authors observed a change in mass corresponding to the loss of four protons from substrate. This product also had absorbance spectral features that suggested formation of a double bonds. When these reactions were treated with iodoacetamide, which alkylates, or adds carbons to, free thiols (e.g. the thiol in Cys), the authors also saw the expected mass shift, which suggests that modified peptide still has a reactive sulfur (Figure 3A).
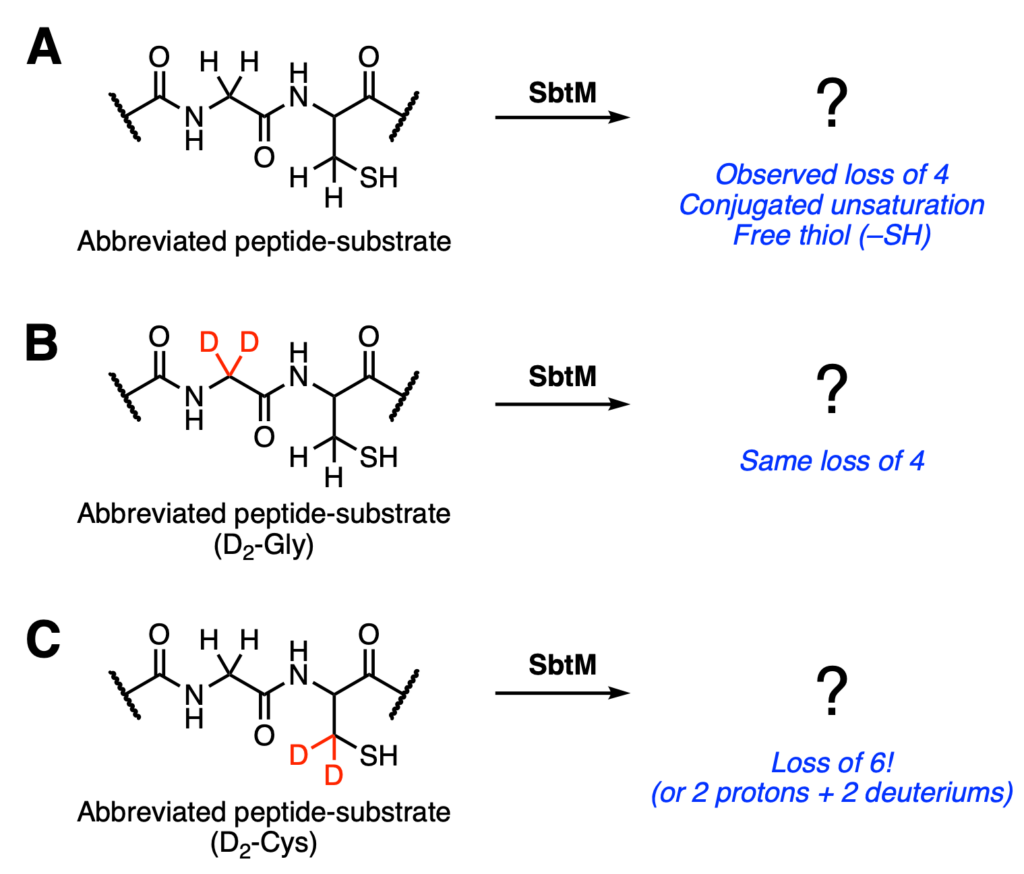
To pinpoint the exact residues involved in this modification, the authors further fragmented the product peptide to identify which residues were affected. Ultimately, the site of modification was traced back to residues Gly34 and Cys35. Indeed, when peptides were synthesized substituting the Cys with Ala or Ser, no reaction was observed, indicating the reactive thiol is important.
Next, the authors tested isotopically-labeled versions of the substrate, where specific hydrogens were replaced with deuterium atoms. In these cases, they used either di-deuterated Gly or Cys. Deuterium is one mass unit heavier than hydrogen. As a result, if you lose a deuterium from the substrate, you will see a mass difference of two rather than one. When the labeled glycine analog was incubated with SbtM, only the expected loss of four was observed (Figure 3B). However, the labeled cysteine analog had a loss of six, which corresponds to the loss of two protons and two deuteriums. Thus, the two deuterated sites are removed from the peptide during the reaction with SbtM. (Figure 3C).
In addition to the prior LC-MS and spectral information, the authors also used nuclear magnetic resonance (NMR) and Fourier-transform infrared (FTIR) spectroscopy to identify changes in the product relative to the substrate peptide. NMR spectroscopy of the product showed a loss in signal corresponding to the Cys amide proton, thus identifying the third proton that is removed from substrate. In addition, there were new features corresponding to an aromatic functional group. FTIR spectroscopy of the product identified changes indicative of changes in the peptide backbone similar to those observed in proline residues. By putting together these individual pieces, the authors believe that SbtM likely catalyzes the formation of an aromatic thiooxazole (Figure 4).
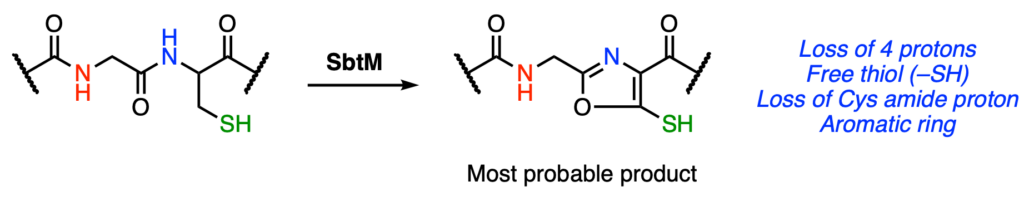
In this work, a variety of biochemical and analytical chemical techniques were used to characterize a new radical SAM enzyme, SbtM. Although not a single experiment could conclusively determine the changes made to the substrate peptide, their collective evidence suggested the formation of a thiooxazole. Additional experiments will hopefully further validate the identity of this new product and explore its functional role in nature. By expanding the known chemistries of radical SAM enzymes, additional natural products with interesting chemical properties will be more easily identified.

