Title: Detection of SARS-CoV-2 RNA Using a DNA Aptamer Mimic of Green Fluorescent Protein
Authors: Bria S. VarnBuhler, Jared Moon, Sourav Kumar Dey, Jiahui Wu, Samie R. Jaffrey
Journal: ACS Chemical Biology
Year: 2022
Featured image by Dwight Sipler is licensed under CC BY 2.0 by Creative Commons.
Whether in a clinical or academic setting, researchers are constantly looking to improve and simplify detection methods. This is no different for the SARS-CoV-2 virus, detection of which normally consists of a PCR-based assay where amplification (or lack thereof) of viral DNA is used to determine if a patient is positive for the virus. However, these tests require a large amount resources and time, often resulting in a testing bottleneck, especially when coronavirus transmission rates are high. Conversely, rapid tests provide results much faster and are more readily available but are not as reliable. In this study, VarnBuhler et al. develop a different kind of detection method for SARS-CoV-2 using a DNA aptamer to address these issues with current tests on the market.
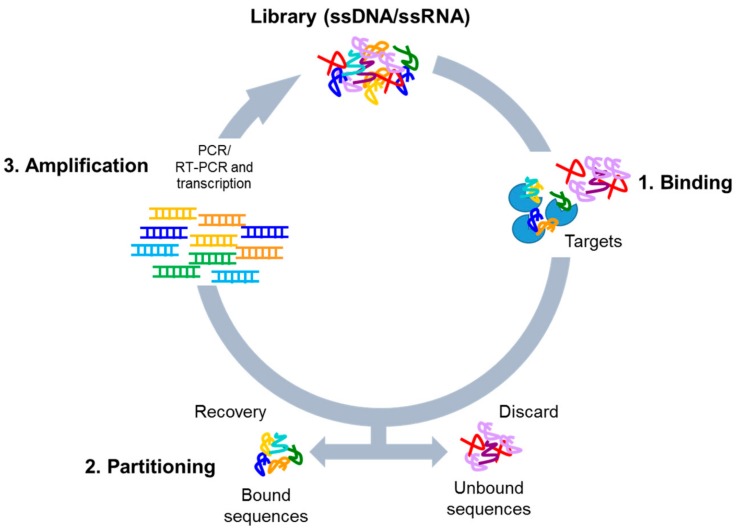
The authors developed a DNA aptamer, called Lettuce, which binds specifically to the fluorophore DFHBI-1T using the systematic evolution of ligands by exponential enrichment (SELEX) technique (Figure 1).1 In this method, libraries of randomized DNA are constructed and subjected to rounds of both “negative” and “positive” selection. During negative selection, the pools of aptamer candidates were incubated with agarose beads linked to an aminohexyl group, designed to mimic the shape of DFHBI-1T.2 Thus, any DNA that binds at this stage does not bind specifically to the fluorophore and is eliminated. Next, the same technique is used in positive selection, except with agarose beads linked to the true fluorophore. Now, DNA binding indicates the aptamer is a possible candidate as it shows affinity for the intended target. This process is repeated to remove DNA sequences which bind nonspecifically, as well as remove DNA sequences which don’t bind well to DFHBI-1T, effectively whittling down the target pool until only a small number of DNA sequences remain. After nine rounds of selection, the researchers identified individual aptamer candidates through bacterial selection and DNA isolation before incubating with the fluorophore.2 Upon addition of DFHBI-1T, the samples with the highest fluorescence output can be sequenced for further testing.
Previous research shows that “splitting” the Spinach RNA aptamer is a useful tool for inducing fluorescence (via binding to DFHBI) only upon additional binding to specific nucleic acid sequences.3 In this technique, the aptamer is split in half, inhibiting the ability of the RNA aptamer to bind the fluorophore and eliminating fluorescence. However, a sequence complementary to the target DNA is added to each aptamer half. Thus, upon binding of each aptamer half to their complementary DNA sequences, which are in close proximity, the two halves of the aptamer are put back together. This restores its ability to bind the fluorophore, but only upon correct binding to the target DNAs.
To apply this functionality to detection of SARS-CoV-2, VarnBuhler et al. designed a split-aptamer system using Lettuce to bind viral RNA (Figure 2).2 To optimize binding efficiency and aptamer functionality, the authors tested multiple binding sites on the SARS-CoV-2 viral RNA, varying parameters such as sequence length and distance between the complementary sequences. Positive hits, indicated by fluorescence upon incubation with viral RNA, support that this technique is effectively able to detect viral RNA. Importantly, the authors also show that the aptamer remains selective for SARS-CoV-2 RNA, as incubation with an unrelated RNA did not induce fluorescence. Additionally, incubation with multiple of the successful aptamer sensors increased fluorescence, with four sensors able to reliably detect viral RNA in concentrations as low as 10 nM.2
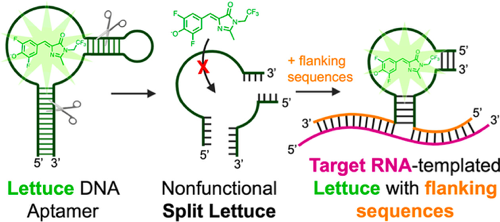
Figure 2. Lettuce is a DNA aptamer selective to DFHBI-1T, a fluorophore. Upon binding, fluorescence is emitted. This technology is used to develop an aptamer which fluoresces only upon binding of SARS-CoV-2 viral RNA. Reprinted with permission from VarnBuhler, B. S.; Moon, J.; Dey, S. K.; Wu, J.; Jaffrey, S. R. Detection of SARS-CoV-2 RNA Using a DNA Aptamer Mimic of Green Fluorescent Protein. ACS Chem. Biol. 2022, 17 (4), 840–853. Copyright 2022 American Chemical Society.
In conclusion, VarnBuhler et al. apply a split aptamer approach to SARS-CoV-2 detection. With the continued prevalence of Covid-19, development of new and improved detection methods of the virus is an important step forward in simplifying diagnosis and treatment.
References
(1) Catuogno, S.; Esposito, C. L. Aptamer Cell-Based Selection: Overview and Advances. Biomedicines. 2017, 5 (3), 49.
(2) VarnBuhler, B. S.; Moon, J.; Dey, S. K.; Wu, J.; Jaffrey, S. R. Detection of SARS-CoV-2 RNA Using a DNA Aptamer Mimic of Green Fluorescent Protein. ACS Chem. Biol. 2022, 17 (4), 840–853.
(3) Kikuchi, N.; Kolpashchikov, D. M. Split Spinach Aptamer for Highly Selective Recognition of DNA and RNA at Ambient Temperatures. ChemBioChem 2016, 1589–1592.

