Title: Resurrected Rubisco suggests uniform carbon isotope signatures over geologic time
Authors: Mateusz Kędzior, Amanda K. Garcia, Meng Li, Arnaud Taton, Zachary R. Adam, Jodi N. Young, Betül Kaçar
Journal: Cell Reports
Year: 2022
Evolution occurs on multiple scales. We are familiar with evolution on the scale of animals. Horses evolved hooves to run long distances; ducks evolved webbed feet to swim; birds evolved wings to fly and so on. However, evolution also happens on the tiniest cells (yeast, bacteria etc.) and even on enzymes. Enzymes, composed of long strings of amino acids, change with time to become more efficient or to specialize their catalytic reaction. A great example of this is the enzyme in photosynthesis that converts carbon dioxide into sugar, called RuBisCo. RuBisCo is thought to be the most abundant enzyme on Earth, because every organism that does photosynthesis and makes oxygen — land plants, phytoplankton, etc. — produces it. Billions of years ago, the ancestor of these all these organisms created RuBisCo for the first time and started to evolve. It began as a marine organism but evolved to survive on land and populate nearly every ecosystem on Earth. This organism is the ancestor of every tree, grass and house plant you see today. During the long evolution of photosynthetic organisms, the RuBisCo enzyme was evolving as well. While geoscientists studying Earth’s history often assume RuBisCo behaved the same way it does today, RuBisCo has mutated significantly through evolution. The authors of this recent study decided to test whether the modern RuBisCo behaves similarly to its ancient ancestor.
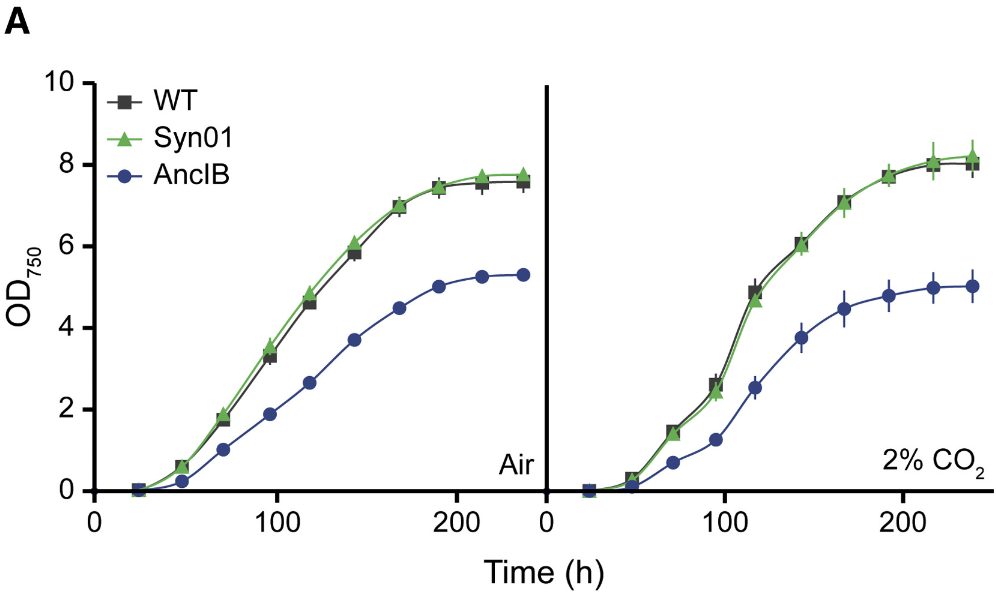
Enzymes can be resurrected from the past by collecting and comparing their descendants and tracing their original amino acid sequence. Each enzyme descendant will have its own variations but should retain most of the original protein’s sequence. By cross-referencing the similarities between RuBisCo descendants in plants, algae, and marine phytoplankton, the authors found the ancestor’s sequence. Then, they synthesized the ancestor DNA code for the enzyme in the lab and inserted it into the genome of a modern photosynthetic organism called a cyanobacteria (Figure 1). They believe this enzyme represents RuBisCo from between 1 and 3 billion years ago. When they grew this ‘ancient’ cyanobacteria strain and compared it with the original strain, they found that the ancient strain could not grow as well. It seemed to make about half as many cells as the original
The authors demonstrated that the inefficient growth of the ancient strain was due to the ancestral RuBisCo. It was possible that the genetic manipulation of the ancient strain made it less efficient. To test this hypothesis, the authors deleted and then reinserted the original RuBisCo DNA into the cyanobacteria. This strain called Syn01, which is genetically identical to the modern but had undergone the same treatment as the ancestral strain, grew similarly to the modern strain. Thus, the genetic experiments were not at fault for the ancestral strains deficiencies. The ancestral RuBisCo simply wasn’t as efficient as the modern form. The ancestral cells tried to compensate by making many, many RuBisCo enzymes per cell, but they still could not keep up. The authors clearly show that the ancestral RuBisCo did not behave in the same way as the modern RuBisCo, answering their initial question.
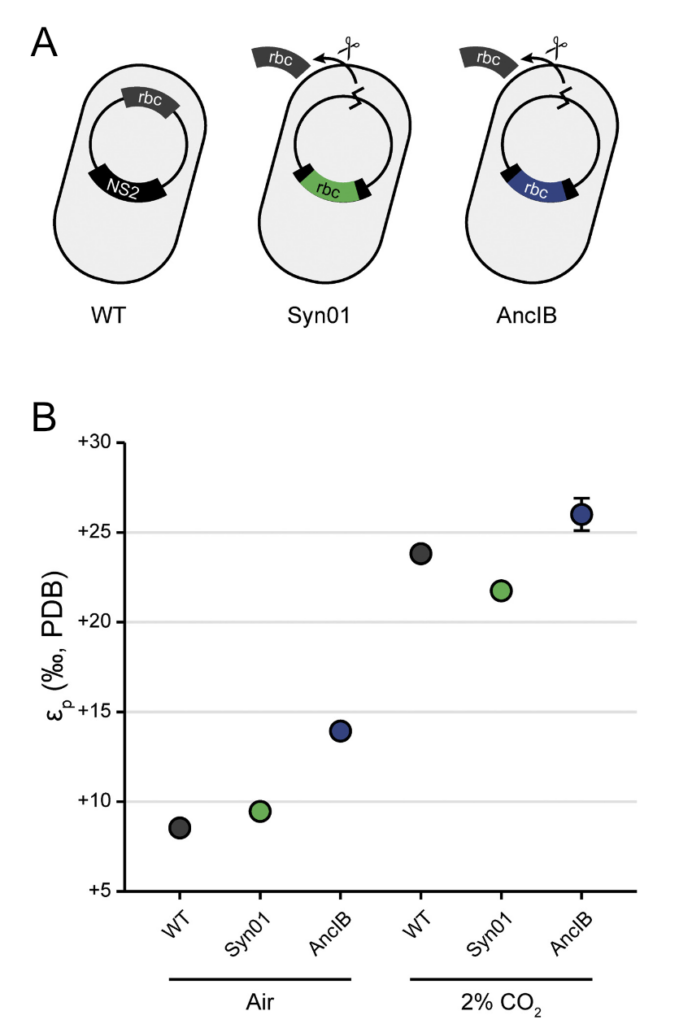
Figure 2: A.) Scheme of genetic edits to the modern (WT), control (Syn01) and ancient (AncIB) strains of the cyanobacteria. B.) The isotope composition of cyanobacterial cells produced by the three strains. They find the ancestral strain produces less 13C than the others. From Kedzior et al. 202
Another open question about the ancestral RuBisCo was whether the sugars (and cell biomass) it created had the same 13C isotope composition as biomass made by modern RuBisCo. Geobiologists often use the 13C composition of buried biomass from billions of years ago to understand how active photosynthesis was through Earth history. If there is more 13C in the ancient biomass, it means more photosynthesis happened and the Earth’s biome as a whole was more productive. However, if the ancestral RuBisCo naturally created cells with more 13C, then you could get the same isotopic signals in buried biomass without needing more photosynthesis. In Figure 2, the value εp records the amount of 13C, where higher values mean less 13C. These results suggest that the ancient RuBisCo made cells with less 13C than modern cells. However, they determined that this difference in 13C composition of the cells was due to the different growth rates of the strains. If the modern strain is grown at the ancestral strain’s slower growth rate, the strains would produce cells with the same 13C composition. Thus, the difference in isotopic compositions was not caused the ancient RuBisCo enzyme itself and the isotopic rock record remains intact.
By resurrecting an ancient enzyme using its modern descendants, the authors of this study were able to directly observe what biological processes looked like billions of years ago. In the case of RuBisCo, these experiments demonstrated that the enzyme was possibly less efficient 1-3 billion years ago, but the isotope composition created by the ancestor was roughly the same as modern organisms. This will make many geobiologists feel more confident in their interpretation of the Earth’s 13C isotopic record. Without a doubt, the resurrection enzymes applied to questions of deep-time evolution will broadly increase our understanding of how life on Earth has evolved. That is perhaps the most lasting impact of this study.

