Article Title: Identification of gut bacterial enzymes for keto-reductive metabolism of xenobiotics
Authors: Qiang, L.; Ouyang, H.; Gordils-Valentin, L. et al.
Journal: ACS Chem. Biol.
Year: 2022
DOI: /10.1021/acschembio.2c00312
We live in a microbial world. Microbes play intricate roles in our daily lives by fermenting foods we consume and fixing N2 for crops to grow. Our own bodies harbor trillions of bacteria within our gastrointestinal tract, which are collectively known as the gut microbiome. These gut bacteria interface with our bodies and can alter xenobiotics (foreign molecules) we consume, including drugs. Identifying which bacteria and which enzymes can catalyze these transformations has large implications on drug development, as these modifications can change bioavailability of drugs in the gut. For instance, the pharmaceuticals nabumetone (1), hydrocortisone (3), and tacrolimus (5) can under ketone reduction by unknown gut enzymes (Figure 1). Here, Qiang et al. report the discovery of such keto-reductases capable of reducing a ketone to a hydroxyl group functional group.
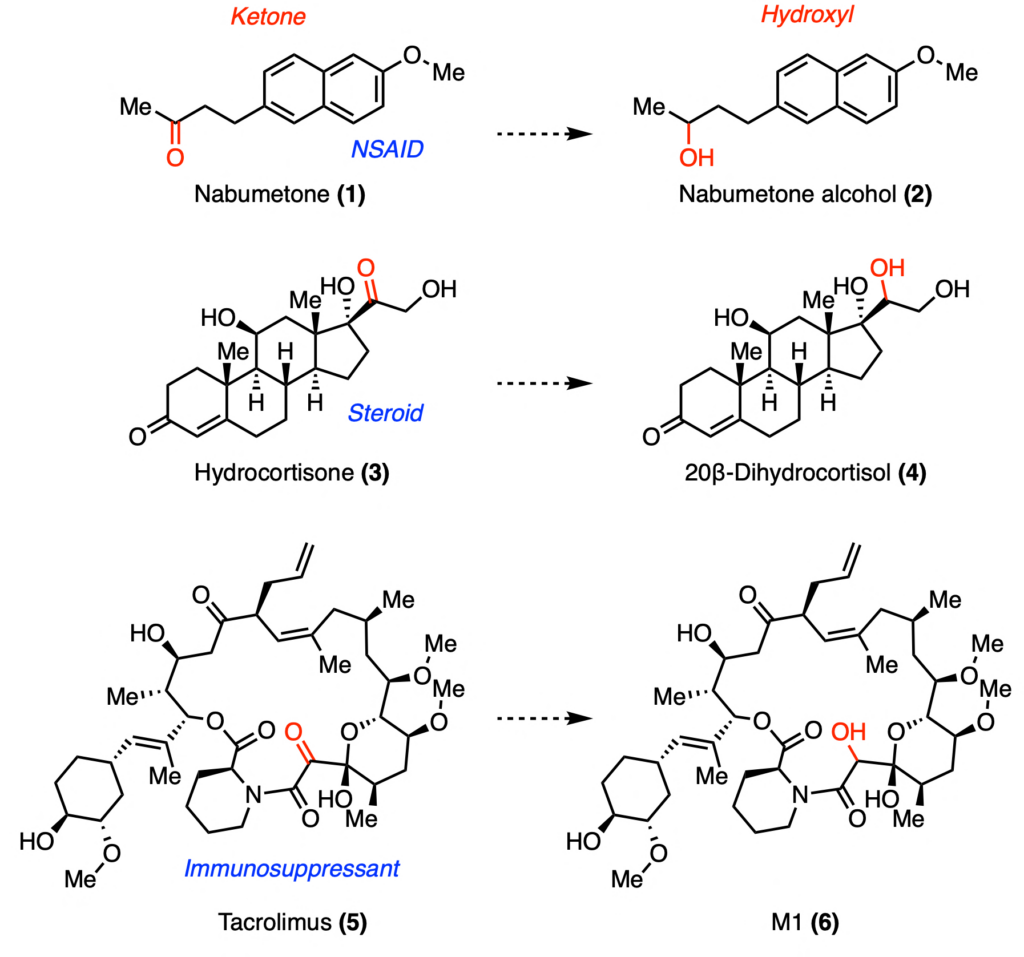
In order to discover candidate ketoreductases, the authors searched within the genome of Clostridium bolteae, a bacterium known to catalyze ketone reduction of xenobiotics. Using a characterized ketoreductase as the query, they identified six enzyme candidates: DesE, DesC, SDR5, TacA, TacB, and SDR15. None of these enzymes had been previously characterized. As such, the authors expressed these enzymes in the model organism Escherichia coli and obtained pure enzymes. These six enzymes were then tested to see if they could convert compounds 1, 3, or 5 into compounds 2, 4, or 6, respectively. By comparing to authentic standards, the authors found that DesE was able to fully metabolize nabumetone (1) and some hydrocortisone (3). In addition, both TacA and TacB were able to partially convert tacrolimus (5).
Although it was great that ketoreductases in C. bolteae were identified, the authors wanted to find enzymes with the same activity in different bacterial species. The composition of every individual’s microbiome is different, so diverse bacteria need to be captured. As a proof of concept, the authors used the newly identified DesE as the input for finding the analogous DesE in E. coli, as E. coli has been reported to act on compounds 1 and 3. Once again, they identified 3 candidate genes (kduD, fabG, yohF), expressed and purified them, and tested them for activity. These experiments showed that FabG could reduce compound 1 but not 3, whereas KduD could reduce compound 3 but not 1.
Due to the successful identification of related DesE ketoreductases in E. coli, the authors set out to identify similar enzymes from human gut microbes. In the end, they were able to find 93 candidates across 40 human gut microbes. Three of these (Ruminococcus torques DesE_R, Blautia obeum DesE_B, and Dorea formicigenerans, DesE_D) were tested alongside the previously discovered enzymes to test the substrate scope of these enzymes. As there are many metabolites within the gut that possess a ketone group, they assessed how structurally diverse potential substrates can be.
A small panel of drugs (Figure 2A) was tested, where those in Group I are simpler and those in Group II are more complex. In total, six enzymes were tested for activity, and the percent conversion is reported in Figure 2B. Based on these data, it appears that only compounds in Group I were metabolized. In addition, each enzyme has a slightly different preferred substrate scope.

In this work, a variety of ketoreductase enzymes from gut microbes were identified and tested for activity in vitro. These enzymes, although related, appear to metabolize different xenobiotics and potentially alter their efficacy or mode of action. With the discovery of these enzymes, additional follow-up studies can be conducted to assess if these enzymes can be found in actual human gut microbiome samples. Just because the gene is present does not mean the enzyme is being expressed and is active. If so, it would be interesting to see if these microbial enzymes can be inhibited with small molecules to prolong the efficacy of various pharmaceuticals.