Authors: Khomkrit Sappakhaw, Krittapas Jantarug, Sarah A. Slavoff, Nipan Israsena, and Chayasith Uttamapinant
Journal: Angewandte Chemie (2020)
Proteins are the hardware of life, comprising a diverse set of shapes and functions to keep organisms alive. The properties of a protein depend on the amino acids that react and fold together to create the overall protein structure. As with any type of hardware, a system runs into issues if it is not functioning properly. In the biological world, many diseases are caused by proteins not behaving like they should.
For example, Alzheimer’s disease (AD) is caused by the unnatural build-up of proteins that form larger plaques and wreak havoc on brain function and synapse communication. Although AD has been studied for many decades, it remains elusive exactly how these peptides form in vivo. More effective treatments are needed for the 25 million patients diagnosed worldwide. To better understand how these plaques form in vivo, researchers from VISTEC and Yale created a method to monitor them using fluorescence.
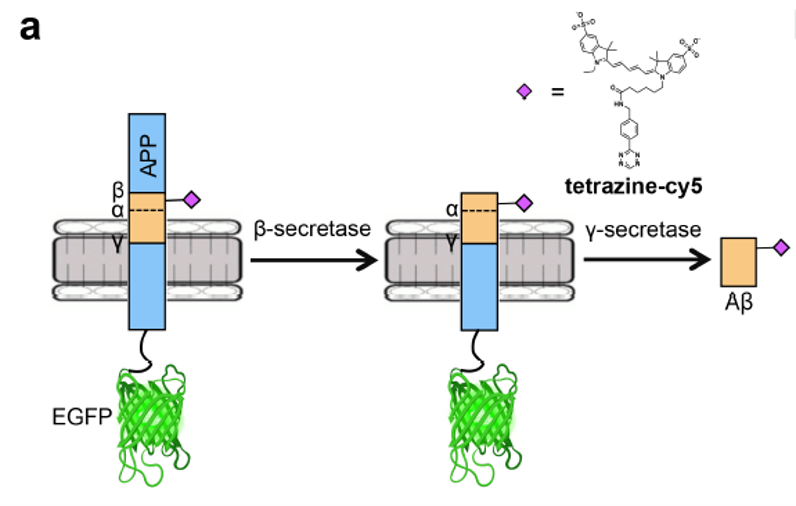
Scientists believe Alzheimer’s disease is caused by the abnormal build-up of proteins in and around brain cells. One of these proteins, amyloid-β, is cleaved from a precursor protein called APP and aggregates into plaques, eventually causing neuron death (Figure 2). In order to visualize these proteins in vivo, the authors found a way to tag both the amyloid-β and the larger APP with fluorescent tags to determine how and where they form in a cell. One of the tags, green fluorescent protein (GFP), is very common. The real trick was incorporating an unnatural amino acid into the amyloid-β protein sequence through genetic code expansion to react with a small fluorescent tag known as tetrazine-cy5.
What is genetic code expansion you might ask? The normal genetic code is a set of DNA sequences that code for the corresponding protein sequence; think software coding for its own hardware. Expanding the code allows organisms to incorporate non-natural amino acids into the proteins it makes. These non-natural amino acids have different properties than their natural counterparts so there are many exciting applications in a variety of fields, including diagnostics and catalysis.
To expand the code, the researchers added a previously engineered piece of cellular machinery, a protein called Methanosarcina mazei pyrrolysyl-tRNA synthetase. This allowed the cells to incorporate the non-natural amino acid bicyclonoyne-lysine (BCNK) into its protein sequences using the RNA code “UAG.”
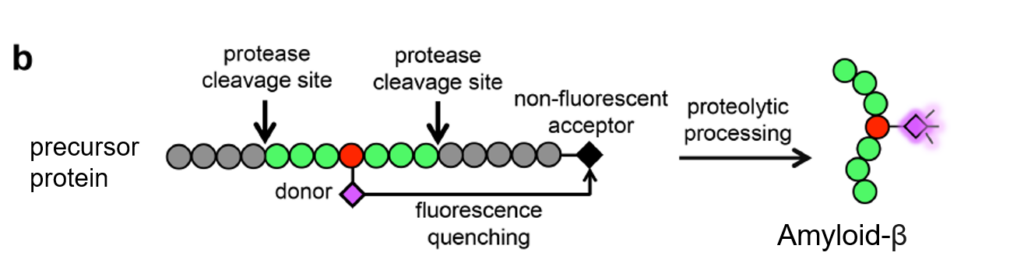
Once the BCNK integrated into the APP protein sequence, the researchers performed a common organic chemistry reaction called Diels-Alder cycloaddition to attach the fluorescent tag tetrazine-cy5 to the unnatural amino acid (Figure 1). Since there are two cleavage steps to produce amyloid-β, two dyes are necessary to figure out which step has occurred. In the first step, an enzyme called β-secretase chops off the top part of APP, leaving an exposed end of amyloid-β. This chopped off portion has no dye and therefore cannot ever be seen. If the second cleavage occurs, the amyloid-β peptide is fully liberated from the GFP-containing APP and fluoresces purple on its own (Figure 2).
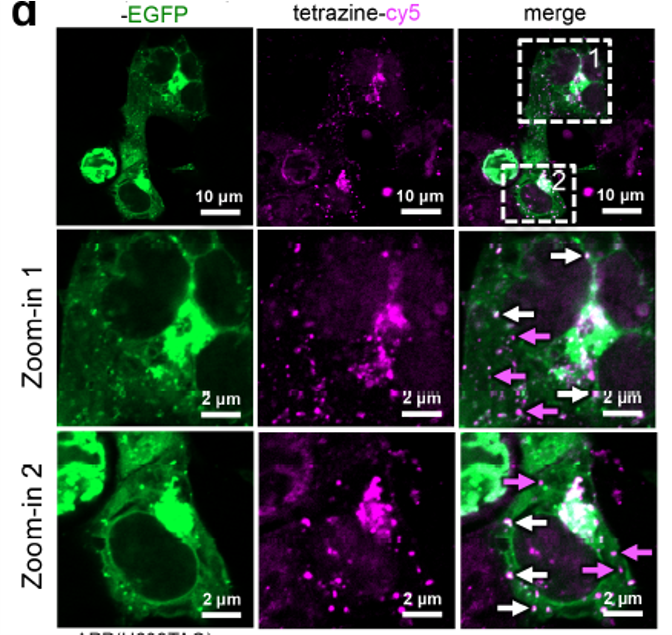
By monitoring both the green fluorescent protein and the BCNK-tetrazine-c5 probe, the researchers could determine when and where cleavage occurred (Figure 3). The purple arrows on the right side of Figure 2 show where the pesky peptide amyloid-β was cleaved and formed plaque-like bunches known as punctas (dyed purple). Purple arrows also point to small dots of the tetrazine-c5 purple dye, which are fully liberated amyloid-β peptides. White arrows point to areas without cleavage, indicated by the presence of overlapped green and purple dyes. As amyloid-β is notoriously difficult to tag, this study presents a significant achievement: the first high-resolution, temporal imaging of APP cleavage.
The author’s results are extremely exciting for the study of Alzheimer’s and related diseases. The unnatural amino acid probe they developed allows high-resolution visualization of amyloid-β formation with potential application to evaluate molecules that prevent the formation of larger plaques. Although it would be difficult to use this probe in vivo with a living human patient, it does bring researchers one step closer to finding a treatment for this debilitating disease.

