Feature photo by Regina Easley from Pixabay
| Title | Selective Neutral pH Inhibitor of Cathepsin B Designed Based on Cleavage Preferences at Cytosolic and Lysosomal pH Conditions |
| Authors | Yoon, M.C.; Solania, A.; Jian, Z.; Christy, M.P.; Podvin, S.; Mosier, C.; Lietz, C. B.; Ito, G.; Gerwick, W.H.; Wolan, D.W.; Hook, G.; O’Donoghue, A.J.; Hook, V. |
| Journal | ACS Chemical Biology |
| Year | 2021 |
| Link | https://pubs.acs.org/doi/10.1021/acschembio.1c00138 |
While many think of disease as a result of a foreign invaders disturbing our normal biological functions (e.g. COVID-19 from the SARS-CoV-2 virus), some arise from transporting our body’s natural material elsewhere. One such enzyme, Cathepsin B, when in its normal, acidic environment will cleave certain protein sequences – but these preferences change in a different pH environment. This has led to severe consequences for neurodegenerative diseases where cathepsin B leaks out of its normal habitat in lysosomes into the cell’s main cavity. Chemists, however, took advantage of this change to design pH-selective inhibitors, preventing cathepsin B’s cancerous activity without interfering with its normal function.
The human enzyme cathepsin B chops up or cleaves discarded proteins in our cells, contained in small, acidic packages called lysosomes. These “recycling centers” gather up proteins and other debris as they reach the end of their utility and use enzymes like cathepsin B to digest them all the way down to their starting materials – spitting out building blocks that the cell can now reutilize for newer structures. But when lysosomes start leaking, as seen in various neurodegenerative conditions like Alzheimer’s disease or stroke, cathepsin B enters the central part of the cell, called the cytosol.
Normally this isn’t an issue – the enzymes contained in lysosomes are only active at the acidic pH, close to that of tomatoes or coffee. However, cathepsin B seems to be an exception; when leaked into the cell’s cytoplasm, at a neutral pH like water, the enzyme continues to break down proteins indiscriminately. This difference in pH was thought to change where cathepsin B cleaves proteins, perhaps leading to its continued activity when other lysosomal enzymes stopped functioning, but no one had proven it yet.
Enter Drs. Vivian Hook and Anthony O’Donoghue (and their students) at the University of California San Diego (UCSD), in collaboration with Dr. Dennis Wolan’s lab at the Scripps Research Institute. Hook is a distinguished professor at UCSD who investigates and creates therapeutic treatments for brain disorders such as Huntington’s disease and traumatic brain injury. O’Donoghue is a master of the Multiplex Substrate Profiling by Mass Spectrometry (MSP-MS) method to figure out the targets of protein-degrading enzymes like cathepsin B in diseases. Together, these scientists defined the details of cathepsin B’s pH-dependent activity, and then used that knowledge to design pH-selective inhibitors and stop its cancerous activity.
In many ways, enzymes are like us with ice cream; we have our absolute favorite flavors, some we will eat but wouldn’t put in their top 10, and some we absolutely will not touch. Enzymes work the same way – they have certain protein sequences that they will cut every time, some they will cut if in proximity but not efficiently, and some they absolutely will not cut. Using O’Donoghue’s expertise in MSP-MS, the authors put cathepsin B together with a library of peptides of differing sequences and monitored the products that formed (Fig 1). Based on which peptides the enzyme cut and at which pH, they could obtain a large amount of information at once on its sequence preferences.
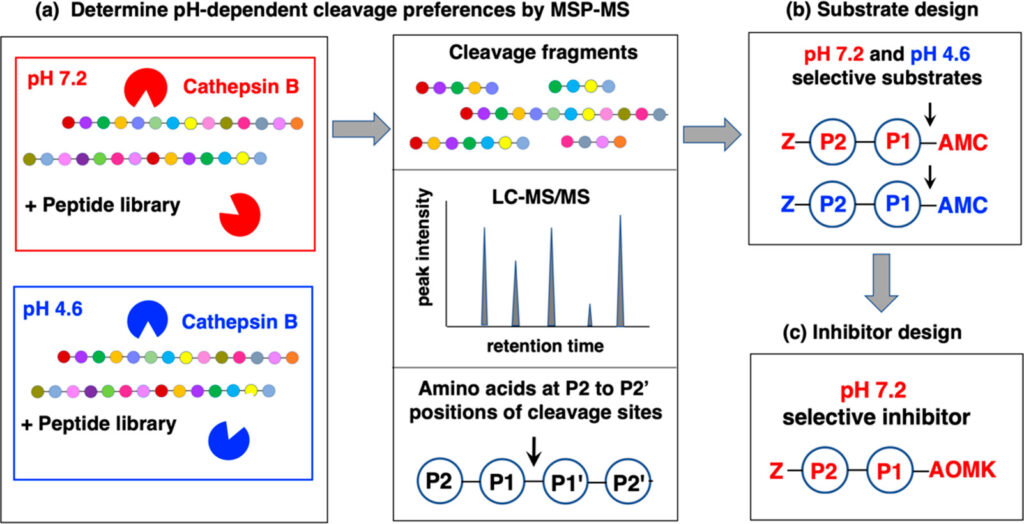
Fig 1. Method process to determine cathepsin B substrate preference and design inhibitors based on their results. Used with permission from Yoon et al. ACS Chem Biol. 2021.
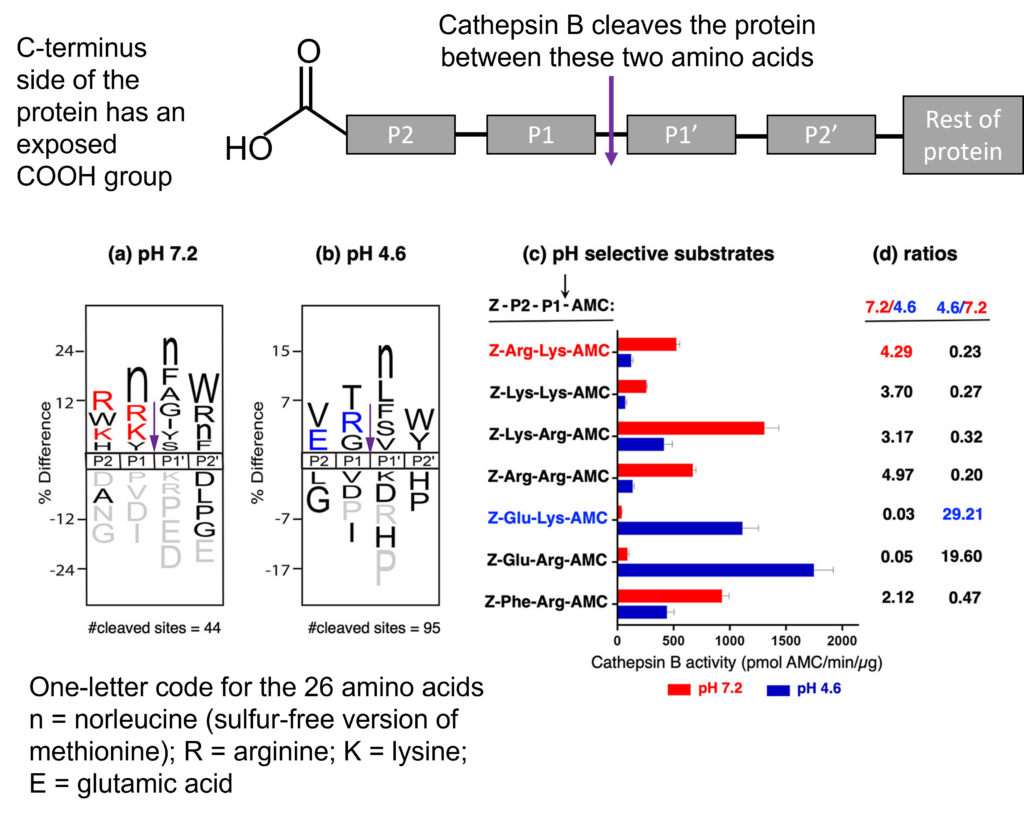
The chemists figured out first that cathepsin B, when in the acidic lysosome, prefers to chop up proteins from the C-terminus end when they have an uncharged glycine (G) or threonine (T) amino acid as the 2nd in the sequence, called the P1 position (Fig 2b). When the enzyme moves into the neutral, cytosolic space, the enzyme prefers to have a negatively charged residue like arginine (R) or lysine (K) in this position (Fig 2a). They further proved this cutting site switch by synthesizing their own peptide sequences with a carboxybenzyl on the N-terminal (denoted as Z) and a 7-amino-4-methylcoumarin (denoted as AMC) reporter on the C-terminal end (Fig 2c). When they had a lysine in the P1 position, they saw significantly more cutting activity from cathepsin B at neutral pH 7.2 than in the acidic 4.6, as evident by the 4x greater ratio of the cut product (Z-Arg-Lys-AMC; Fig 2c) at pH 7.2. Likewise, when glutamic acid was synthesized at the P2 position and a lysine or arginine residue in the P1 position, they saw almost 29x more cut product at the acidic pH than the neutral pH (Z-Glu-Lys-AMC; Z-Glu-Arg-AMC; Fig 2c).
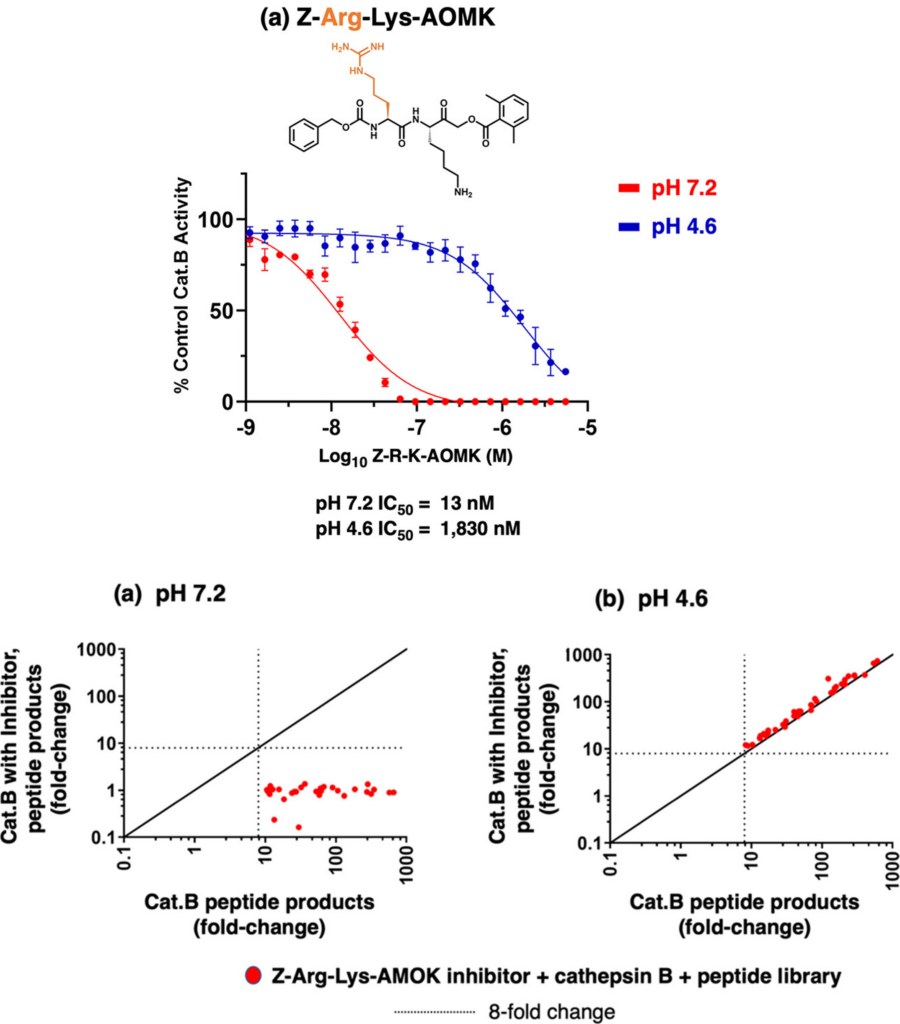
These Z-peptide-AMC compounds revealed how they could entice cathepsin B to interact with their drug at a neutral pH but not at an acidic pH. The scientists then took it a step further and replaced the AMC with a chemical warhead called AOMK. Warheads interact with enzymes via irreversible covalent bonds, occupying the enzyme’s active site and preventing other substrates from being cleaved. The preferred peptide cleavage sequence acts as a bait to lure the enzyme in, then permanently inactivates it by putting their chemical warhead in proximity to the active site. The authors showed that when their most promising Z-Arg-Lys-AOMK drug interacted with cathepsin B, very little inhibitor was required to prevent the enzyme’s function at the neutral pH 7.2 but required almost 100 times as much to inhibit with the same effectiveness at the acidic pH 4.6 (Fig 3 top panel A) – demonstrating the drug’s pH-selective inhibition. Furthermore, when the scientists allowed the Z-Arg-Lys-AOMK compound to mix with cathepsin B before adding it to the peptide library (containing almost 230 different substrates), the enzyme could barely cleave the amount present at the neutral pH but functioned perfectly at the acidic pH (Fig 3 bottom panels A and B).
Yoon et al.’s study on cathepsin B’s activity in different environments led to their creation of a pH-selective inhibitor with promising clinical potential. For those suffering from lysosomal leakage, the ability to inhibit cathepsin B’s activity outside of its natural habitat could ameliorate inflammation and discourage an invasive or pro-metastatic state of various tumors. This paper perfectly shows that by understanding the basic function and preferences of our body, scientists can produce practical drug design and more targeted clinical candidates for therapeutic treatment.