Featured image: Adapted from H. U. Kim, D. G. Choi, H. Lee, M. S. Shim and K. W. Bong, Lab Chip, 2018, 18, 754 with permission from The Royal Society of Chemistry.
Title: Fabrication of dual stimuli-responsive multicompartmental drug carriers for tumor-selective drug release
Authors: Hyeon Ung Kim, Dae Gun Choi, Hyunjee Lee, Min Suk Shim, and Ki Wan Bong
Journal: Lab on a Chip
Year: 2018
https:// dx.doi.org/10.1039/C7LC01063J
In a previous Chembite, we explored a new way to selectively target tumor cells with cancer drugs. Precise delivery methods make sure that the expensive and hard to make cancer drugs are as effective as they can be while also minimizing harmful effects in other parts of the body. But while that method focused on controlling tiny nanomotors that moved on their own to the cancer cells, there are many other ways to focus treatment on only the area that needs to be treated.
One common method is to trap the drug inside of something that only releases it when near a tumor. Cancer cells work differently than most of the cells in our body, which causes the chemical environment around tumors to be different too. Notably, these “tumor microenvironments” have a higher concentration of reducing agents (which can break bonds by adding electrons to a molecule) and are often more acidic than the rest of your body. Because of this, drugs can be trapped inside of structures that break down when triggered by reduction or acidic conditions (low pH). As a result, cancer drugs are only released near the tumor cells. In this paper, researchers have found a way to combine these two “triggers” and have different environments release different drugs from the same pill.
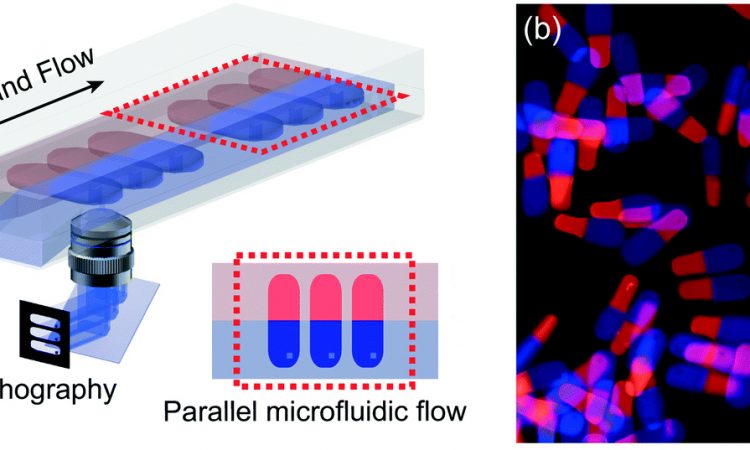
Part of the chemistry behind this customization is polymerization, where long molecules called “polymers” are created by chemically joining many smaller molecules, called “monomers.” This can sometimes be done with high energy ultraviolet (UV) light, as was the case in this paper. When these polymers form, they trap whatever smaller molecules they are mixed with inside the larger structure. But if these monomers have parts that can be broken by certain chemicals (like reducing agents or acids), then the entire polymer can be dissolved and whatever is trapped inside can be released (Figure 1A).
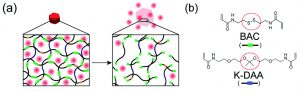
Figure 1: (A) Cartoon of polymer network and breakdown Green portions are the breakable bonds in the monomers and the red dots are trapped and released drugs. (B) The breakable monomers BAC and K-DAA. The breakable bonds are circled in red. Adapted from H. U. Kim, D. G. Choi, H. Lee, M. S. Shim and K. W. Bong, Lab Chip, 2018, 18, 754 with permission from The Royal Society of Chemistry.
These researchers tackled the issue of cancer treatment in exactly this way, using two different polymer mixtures with different chemical properties. One polymer was made with a molecule called N,N’-Bis(acryloyl)cystamine (BAC), that has a disulfide bond (a bond between two sulfur atoms) that can be broken by reducing agents (Figure 1B). The other was made with a different molecule, ketal-bearing diacrylamine (K-DAA). This molecule contains a ketal group, which can be broken by acids (Figure 1B). After making solutions with these monomers and other chemicals that model real cancer drugs, the researchers shine UV light and cause solid polymers to form that trap the model drugs within them. Then, when a reducing agent or acid is added to this, the specific polymer breaks down and releases the model drug.
To actually make the polymer beads, the researchers used a combination of two techniques: microfluidics and stop-flow lithography. Both of these are rich and active fields of research which you can find more about by reading the paper and other Chembites! But briefly, small amounts of liquid are flowed through tiny channels, where UV light is shone on just some areas. The polymer beads are made only where the light hits the solution, and many tiny beads are formed and washed away. This takes less than a second before it all starts again and thousands of beads are made every minute.
The researchers first tested this by making beads with BAC as the only breakable monomer. They trapped fluorescently-labeled proteins inside of the beads as a model for cancer drugs, which can be protein-based. When they added higher concentrations of dithiothreitol (DTT), the beads dissolved faster (Figure 2A), releasing different amounts of the model drug (Figure 2B). This shows that if these beads were in the body, they wouldn’t dissolve in normal tissue (which has a low concentration of reducing agents), but would dissolve and release the drug near tumors (which have high concentrations of reducing agents).
Figure 2: (A) BAC-containing beads in solutions with a reducing agent (DTT). More DTT makes the beads break down. (Scale bar 50 µm) (B) Lower concentrations of DTT make the beads release less model drug. Adapted from H. U. Kim, D. G. Choi, H. Lee, M. S. Shim and K. W. Bong, Lab Chip, 2018, 18, 754 with permission from The Royal Society of Chemistry.
To increase the complexity of this system even further, the researchers decided to create beads that had different compartments. One area contained BAC and a fluorescently-labeled protein. The other contained K-DAA and small fluorescent particles (which modeled a different form of cancer drug). Each solution was flowed in a single channel inside of a microfluidic device, until they reached a place where they could touch each other and flow side-by-side (a process called laminar flow). Then UV light was shone across both liquids, making beads that were half BAC-based and half K-DAA-based (Figure 3A).

Figure 3: (A) Cartoon of double-sided beads forming with stop-flow lithography. (B) With reducing conditions (“Redox”), only the BAC-based side dissolves. With acidic conditions, only the K-DAA side dissolves. With both reducing and acidic conditions, the entire bead dissolves. (Scale bar 50 µm) Adapted from H. U. Kim, D. G. Choi, H. Lee, M. S. Shim and K. W. Bong, Lab Chip, 2018, 18, 754 with permission from The Royal Society of Chemistry.
When put in just a reducing solution, the red side of the bead (with BAC) dissolved, releasing the fluorescently-labeled protein into solution (Figure 3B). But in an acidic solution, just the blue half (with K-DAA) disappeared, allowing the beads to float out. If the solution was both reducing and acidic, the entire bead dissolved and both model drugs were released.
A treatment like this could be used in the future to fine-tune what drugs are released and where they are released within the body. Certain types of tumors can be acidic, but not reducing. And other tumors are reducing, but not acidic. A system such as this could be injected into a tumor and then only release the drug that would be the most effective for that specific type of tumor.
Medical science has progressed at a breakneck pace over the past few centuries and it is only a matter of time until fully personalized treatments are a reality. The research described here is a step in this direction, potentially allowing doctors to design customized drug cocktails for a single patient. No two cancers are exactly the same; it makes sense that our treatments should be just as flexible.