Authors: Shanshan Tian, Ranran Liu, Zhiyue Wu, Kai Zhang
Journal: Analytical Chemistry (ACS)
Year: 2022
Featured image by mutantmommy from Pixabay.
—
You are what you eat – so you might not want to eat illegal chemicals added to your food. Chemical contaminants in foods, from pesticides to preservatives, continue to affect human health and the environment. Despite worldwide food regulation systems banning the illegal addition of prohibited chemicals to foods, consumers are still exposed to unstudied and ill-regulated chemicals. Scientists need sensitive and selective ways to identify these chemicals in food products, even if the food is washed after adding the chemical.
In December 2022, a research team from Tianjin Medical University developed a novel mass spectrometry method for detecting permanent traces of formaldehyde (FA) before and after washing fresh food products (shrimp, fish, pork, and beef).1 Tian et al. found that detecting proteins created by FA’s chemical reactions succeeded as a permanent marker for determining any FA addition in food products, even after rinsing the food. And because the method was reliable for both small and large samples of food, the method has the potential to be widely used in detecting added FA in food safety analyses.
Formaldehyde (FA) is a tricky chemical in food regulation as it is often used to sterilize food products and prevent bacterial growth. The U.S. Food and Drug Administration has provided guidelines for formaldehyde as an indirect food additive – it should not be added in excess. However, though the Internal Agency for Research on Cancer and the U.S. Environmental Protection Agency declared prolonged exposure to FA as likely “carcinogenic to humans” when inhaled or ingested, it continues to be excessively added to foods for its effective preservation properties. Current analytical methods, including spectrophotometry and chromatography, can detect FA in food products but can’t necessarily tell if FA has ever been added to food products that are washed entirely afterward.
Tian et al. wanted an analytical method that was reliable for detecting FA added at any time to foods. Since they knew FA could modify proteins present in the foods, they hypothesized that the more FA was added to food, the more modified proteins would be in the foods. The method successfully detected this trend in modified proteins in a variety of unrinsed and rinsed food products with added FA, showing its unique sensitivity and potential for widespread excess FA detection in food regulation systems.
Because FA is reactive, it can cause chemical reactions with biological molecules. FA has been shown to chemically modify amino acids, the units that make up proteins. By studying the modified proteins with a mass spectrometry technique called proteomics, scientists can determine if the proteins came from FA-caused reactions. Figure 1 shows the steps involved in the proteomics approach for protein modification analysis. Tian’s research team used these steps to develop a unique method for detecting FA in unwashed and washed food products.
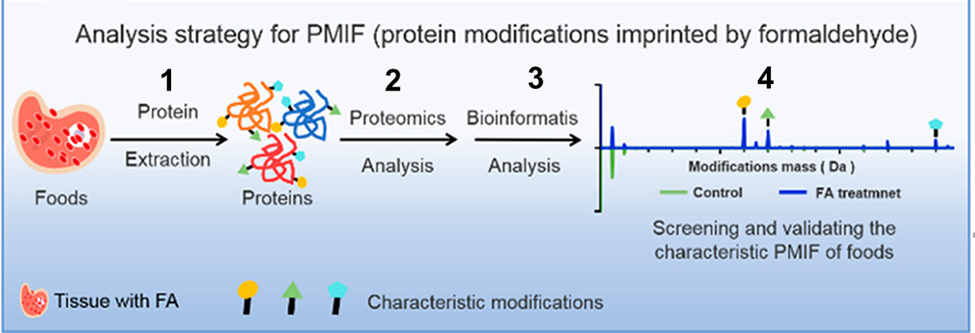
Figure 1. To assess control and FA-treated products, modified proteins are (1) extracted from sample tissues and (2) analyzed by mass spectrometry proteomics. The mass spectra results are (3) assessed by software to (4) identify peaks associated with protein modifications from FA reactions compared to control samples (PMIF = protein modifications imprinted by formaldehyde) (modified from ref. 1).
To test the analytical method with a variety of foods, the scientists took tissue samples from shrimp, pork, beef, and salmon. One group of samples was soaked in water with no FA, while the other group was treated with FA. This allowed them to compare the differences in protein modifications with and without added FA. After extracting the protein from food tissues and breaking down the samples (with tryptic digestion), the research team recorded the ions produced from each sample using mass spectrometry.
The mass spectral results from shrimp tissue showed peptides (smaller subunits of proteins) appear to have four modifications. Figure 2A shows the differences in mass spectra between the control and FA-treated samples that hint at protein modification.
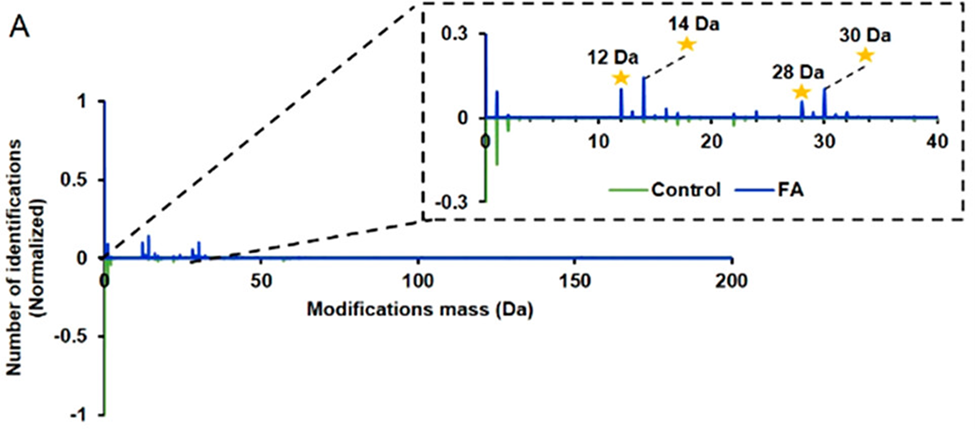
Figure 2A. Comparison of the spectrum from FA-treated samples (blue lines) and control (water-soaked) samples (green lines). The yellow stars represent mass shifts on amino acid residues, indicating protein modifications have occurred (adapted from ref. 1).
But the scientists needed to verify these changes were caused by FA reactions. So, they conducted experiments on three sample levels: peptide, cell, and tissue (from beef). Each sample was treated with FA and mass spectral results were compared to non-treated samples. In all of the tests, they figured out that the same protein modifications were present in these FA-treated samples and the results differed significantly from the control samples.
A significant success – the scientists showed FA caused the protein modifications. Now they needed to prove their mass spectrometry method was reliable for determining how much FA is present in food products. For instance, you can know baking soda and vinegar produce carbon dioxide bubbles, but that doesn’t tell you how many bubbles are produced by a certain combination of baking soda and vinegar. The team treated salmon with FA at various concentrations and soaking times. Two important discoveries: (1) as the amount of added FA and soaking time increased, the mass spectrometry signals from the four protein modifications increased; (2) the method gave consistent results of protein modification signals. These results show that protein modification mass spectral analysis is a trustworthy method for measuring excess FA additions in food products.
Finally, the researchers tested if the same protein modifications could detect FA in products washed after adding FA. After comparing FA-treated samples (salmon, shrimp, beef, and pork) with and without rinsing, they determined the four protein modifications were clearly detected in each sample. Figure 3 shows how the FA-caused protein modifications were detected in both washed and unwashed samples of salmon.
This makes Tian et al.’s technique unique from traditional analytical methods as it can detect permanent evidence of FA in foods, whether the illegally treated food has been washed or not. And with the approach being consistent with all four food products tested in small (1-5 samples) and large samples (>70 samples), the method holds potential for wide use in excess FA detection during food regulation analysis.
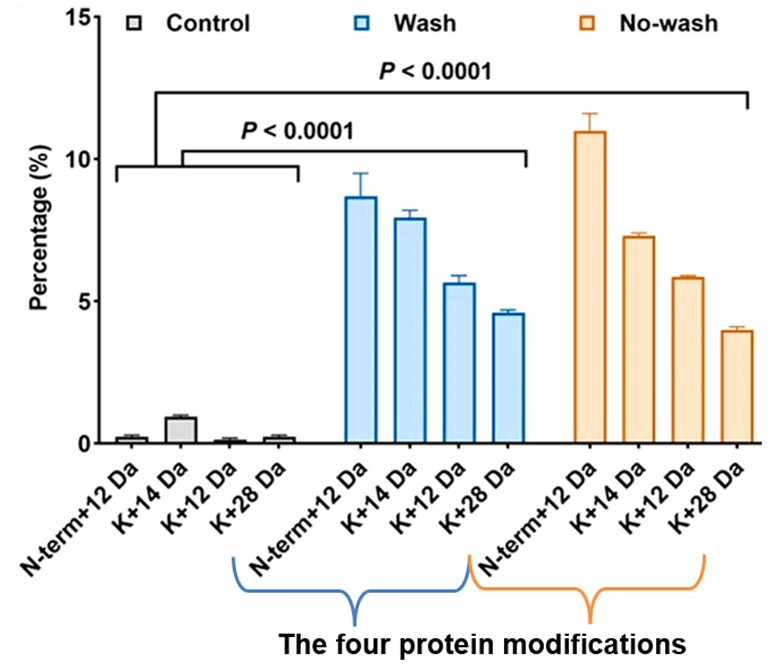
Figure 3. The percentage of each protein modification found in the control (washed, no FA, gray bars), washed (with FA, blue bars), and unwashed (with FA, orange bars) samples. As seen in the blue and orange sections, the modifications from FA were found in both unwashed and washed treated samples. (modified from ref. 1).
Using Tian et al.’s novel protein modification-based mass spectrometry method will determine if FA has ever been illegally added to food products (before and after washing), making it a unique food analysis approach compared to previous analytical methods. With continued development, this method can be applied to other illegal chemical additives and help food safety analysis keep people safe from harmful chemicals in foods.
—
Figures reprinted with permission from: Tian, S., Liu, R., Wu, Z., & Zhang, K. A Novel Strategy Based on Permanent Protein Modifications Induced by Formaldehyde for Food Safety Analysis. 2022. Analytical Chemistry. Copyright 2022. American Chemical Society.
Reference 1: Tian, S., Liu, R., Wu, Z., & Zhang, K. A Novel Strategy Based on Permanent Protein Modifications Induced by Formaldehyde for Food Safety Analysis. Anal. Chem. 2022, 94(50), 17365-17369.
DOI: https://doi.org/10.1021/acs.analchem.2c04069

